Local versus Global Biological Network Alignment
Reference: Lei Meng, Aaron Striegel, Tijana Milenkovic. "Local versus Global Biological Network Alignment," Bioinformatics, in press, 2016.
1. Code and Data Download
You can download our software as either graphical user interface (GUI) or source code for any platform with Python 2.7, SciPy, and wxPython (the latter is needed for GUI only) installed.
To install Python 2.7 and SciPy, simply download and install Anaconda from https://www.continuum.io/downloads. Then, to install wxPython, run in the Anaconda Command Prompt the following command: conda install -y wxpython.
You can download networks from our paper here.
You can download alignment results from our paper here.
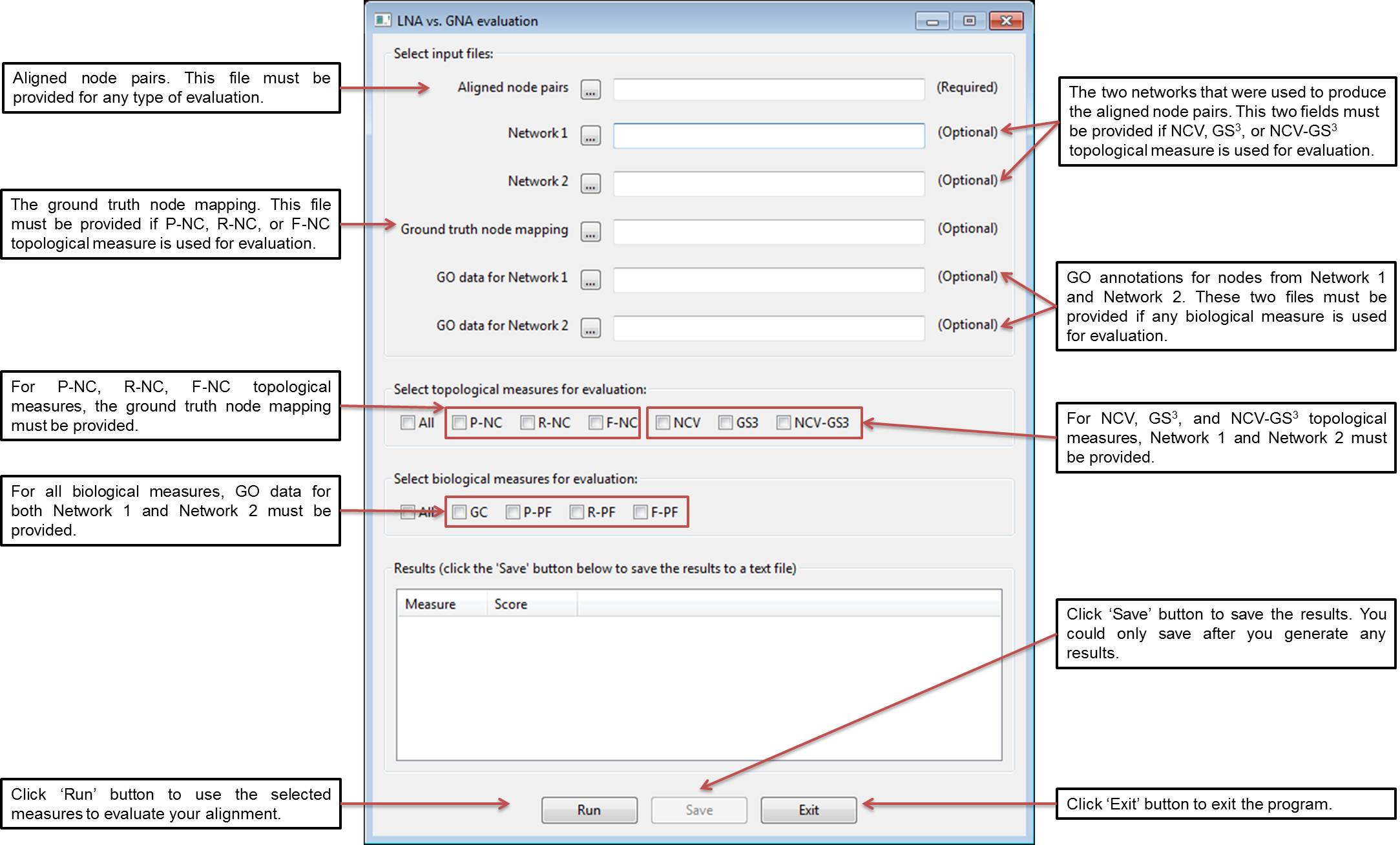
2. Usage Instructions for the Graphical User Interface (GUI)
Start the GUI by running the following command: python GUI.py (note that if you are using the GUI on Mac OS, you might need to run the following command instead: pythonw GUI.py). Below is a screen shot of the GUI interface along with the description of each input field. The format of each input file is described in details in part 3.

3. Usage Instructions for the Python Source Code
The command syntax to run the source code is
python main.py $1 $2
where
$1 is the configuration file. In the configuration file, you need to specify one or more of the options listed in the table below, depending on the alignment quality measures of your interest. See an example.
$2 is the output file that will store the evaluation results after they are computed. The output file contains the results in tab-delimited format (see an example), with the first and second columns denoting the measures used and their corresponding alignment quality scores.
| Option | Comment |
| Aligned node pairs | The alignment whose quality is to be computed, i.e., a file containing the list of aligned node pairs in tab-delimited format (see an example), with the first and second column denoting the nodes from the first and second network, respectively. Two nodes in each line are those nodes from each network that are aligned to each other. Note that some NA methods produce aligned node clusters rather than aligned node pairs, in which case you can convert the aligned node clusters into aligned node pairs by assuming that in a given cluster each node is aligned to every other node in that cluster. This file must always be provided as input. |
| Network1 | The first of the networks being aligned, provided in LEDA (.gw) format. See an example. A description of the LEDA graph format is available here. Note that the header of our network format contains only three lines, which are the same as the first three lines described here. If your network is in edge list format, you can use the script "list2leda" provided here to convert it into the LEDA graph format. This file must be provided if any of the NCV, GS3, and NCV-GS3 topological measures is to be computed for the above input alignment. |
| Network2 | The second of the networks being aligned, provided in LEDA (.gw) format (the same format as for Network1). This file must be provided if any of the NCV, GS3, NCV-GS3 topological measures is computed. |
| Ground truth node mapping | The file containing the list of node pairs that are known to be mapped to each other according to true node mapping (rather than according to the above input alignment), provided in tab-delimited format (see an example), with the first and second column denoting the nodes from the first and second network, respectively. Two nodes in each line are those nodes from each network that are known to be mapped to each other. This file must be provided if any of the P-NC, R-NC, and F-NC topological measures is computed. |
| GO data for Network1 | The file containing the list of gene-GO term annotations for nodes in Network1, with the first and second column denoting the proteins and their assigned GO terms, respectively (see an example). This file must be provided if any of the P-PF, R-PF, F-PF, and GC biological measures is computed. |
| GO data for Network2 | The file containing the list of gene-GO term annotations for nodes in Network2, with the first and second column denoting the proteins and their assigned GO terms, respectively (see an example). This file must be provided if any of the P-PF, R-PF, F-PF, and GC biological measures is computed. |
| Measures | List of desired topological or biological alignment quality measures to be computed on the input alignment, separated by commas, where the alignment quality measures are a subset of: P-NC, R-NC, F-NC, NCV, GS3, NCV-GS3, P-PF, R-PF, F-PF, and GC. For more details about these measures, please refer to our paper. At least one measure must be provided as input. |
4. Links to Implementations of the Existing NA Methods Evaluated in Our Study
Our software is intended to allow for analyzing an alignment that is provided as input into the software. It does not implement the existing NA methods that produce the input alignment in the first place. The latter is certainly of interest, but this is an extremely challenging task. This is because the different NA methods are typically developed by different research groups and are often required to run in different system environments. Thus, it would be very hard (perhaps even not permitted) to integrate them into our software. As an alternative, below we provide links to software implementations of the existing NA methods evaluated in our study. Users can produce a network alignment by running any of the NA methods listed below or any other NA method of their interest, and then input the resulting alignment into our software to evaluate its quality.
| Network aligner | Local or global? | Link to the software |
| NetworkBLAST | Local | http://www.cs.tau.ac.il/~bnet/networkblast.htm |
| NetAligner | Local | http://netaligner.irbbarcelona.org |
| AlignNemo | Local | http://www.bioinformatics.org/alignnemo/wiki/ |
| AlignMCL | Local | http://sites.google.com/site/alignmcl |
| GHOST | Global | http://www.cs.cmu.edu/~ckingsf/software/ghost |
| NETAL | Global | http://bioinf.modares.ac.ir/software/netal |
| GEDEVO | Global | http://gedevo.mpi-inf.mpg.de/ |
| WAVE | Global | http://nd.edu/~cone/WAVE/WAVE.zip |
| MAGNA++ | Global | http://www3.nd.edu/~cone/MAGNA++/ |
| L-GRAAL | Global | http://bio-nets.doc.ic.ac.uk/L-GRAAL/ |