Genetics of Larval Saltwater Tolerance in Anopheline Mosquitoes
Salinity tolerance in the larval stage has arisen repeatedly and recently in several culicid lineages and, according to the working phylogenetic hypothesis for the An. gambiae complex, has arisen at least two times in this group alone (in An. merus, An. melas, and An. bwambae). Outside of the An. gambiae complex, other malaria vectors in Europe, Southeast Asia, Australasia and the Neotropics are often embedded in species complexes whose members have differing larval ecologies and salinity tolerance. Clearly, salinity tolerance plays a key role in determining habitat use and ecological distribution of mosquitoes, and thus their contribution to malaria transmission.
Unlike other complex ecological, behavioral and life history traits of epidemiological importance that are probably polygenic, saltwater tolerance is relatively tractable, likely governed by a few major loci with large effects, and simple to assay. There is an established and growing literature on the physiology of osmotic and ionic regulation in individual mosquito species, especially culicines. Yet little is known regarding the genetic basis of this trait in mosquitoes. The ability to dissect the genetic basis of this adaptive trait using next generation genomic tools lays the groundwork for future efforts to understand the mechanisms by which malaria vector mosquitoes adapt to a heterogeneous and changing environment.
Our long-term goal is to elucidate the genetic basis of salinity tolerance in all three halophilic members of the An. gambiae complex and ultimately in other anopheline species, to determine whether this trait has a common genetic basis. We are dissecting saltwater tolerance through the complementary strategies of quantitative trait locus (QTL) mapping, gene expression profiling, and immunohistochemistry.
Collaborators:
J. Romero-Severson (University of Notre Dame), P. Andolfatto (Princeton Unversity), B. White (University of California, Riverside), P. Linser (Univ. FL Whitney Laboratory).
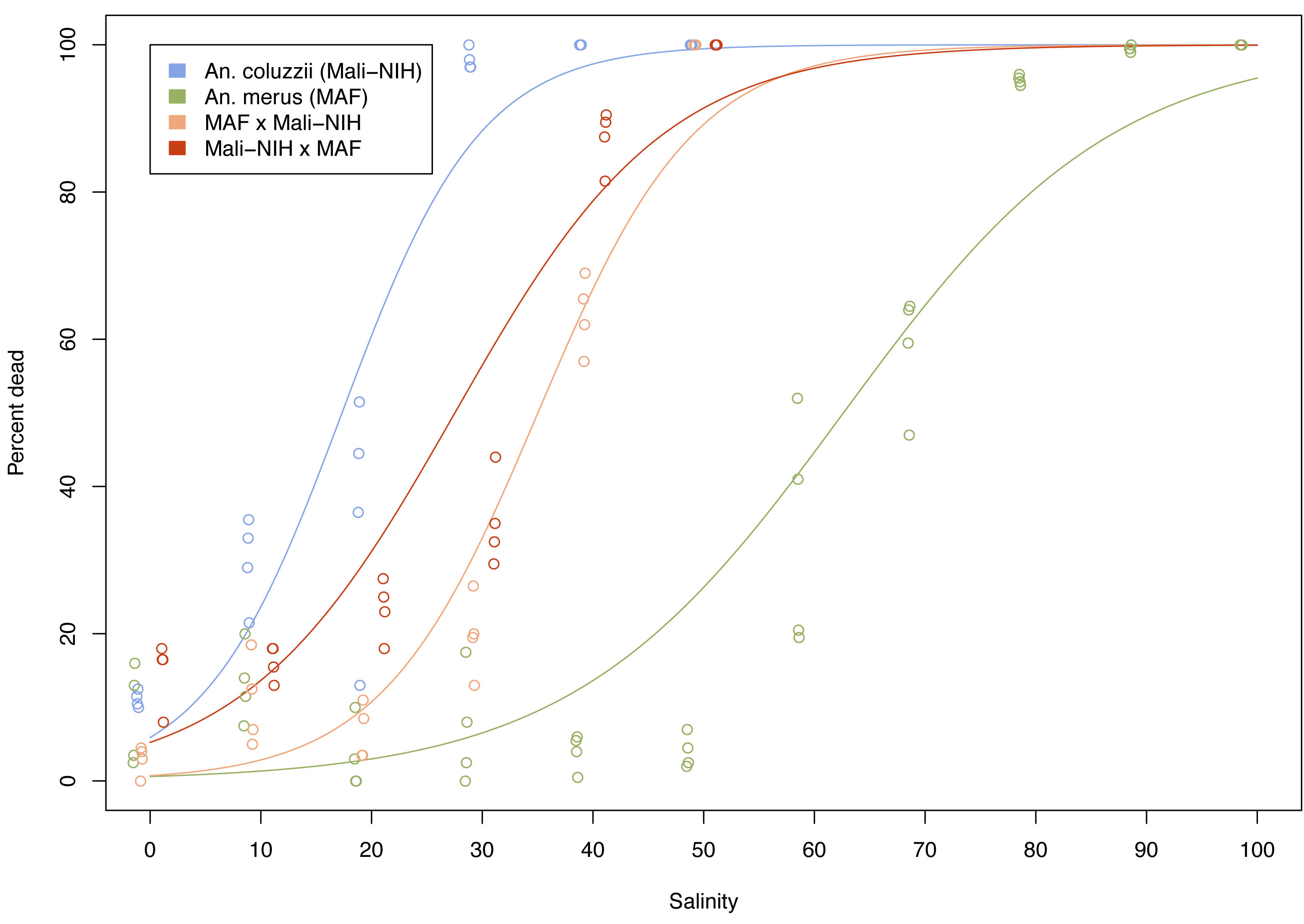 Fig. 1. Dose-mortality response to chronic saltwater exposure in larvae from parental colonies and their F1 hybrids from reciprocal crosses
Fig. 1. Dose-mortality response to chronic saltwater exposure in larvae from parental colonies and their F1 hybrids from reciprocal crosses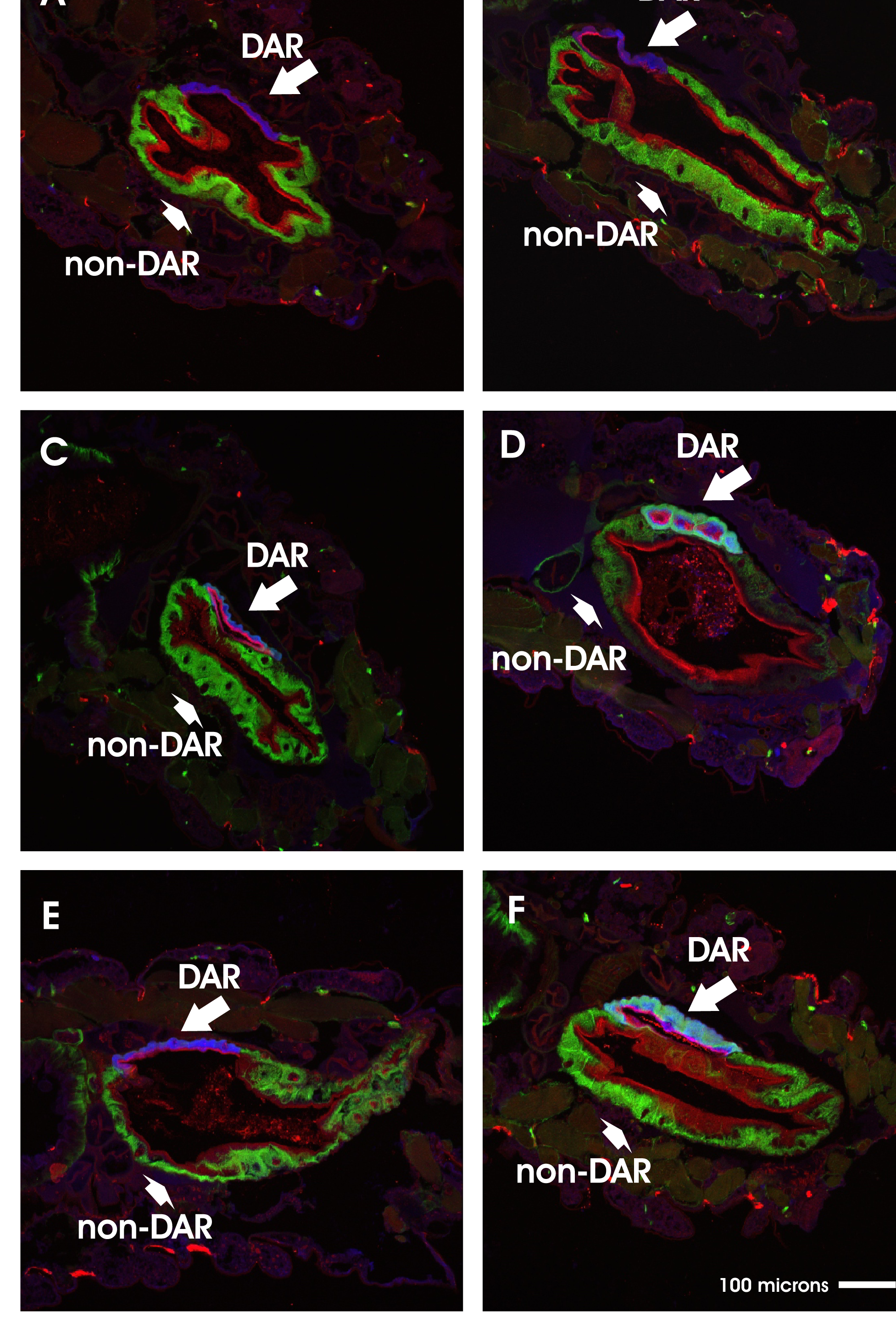 Fig. 2. Comparison of rectal localization patterns of major ion transport proteins in An. coluzzii and An. merus larvae reared in freshwater versus saltwater
Fig. 2. Comparison of rectal localization patterns of major ion transport proteins in An. coluzzii and An. merus larvae reared in freshwater versus saltwater