Evolutionary Genomics of the Y Chromosome in An. gambiae
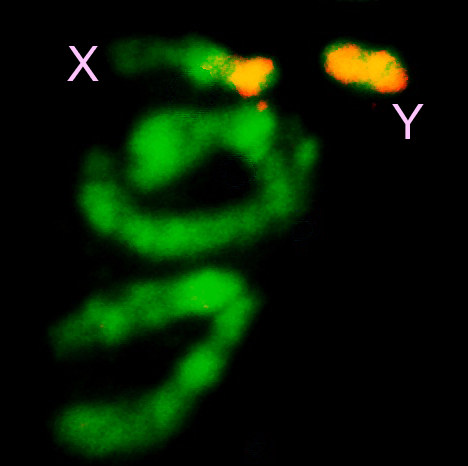
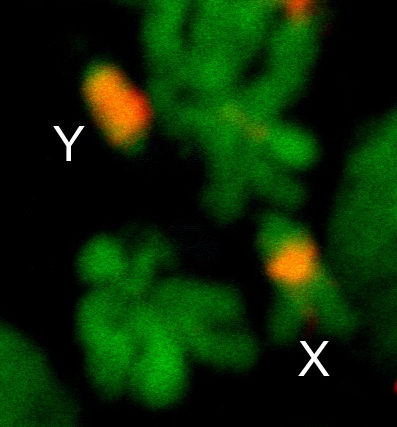
In organisms that have heteromorphic sex chromosomes, such as Anopheles mosquitoes and Drosophila, the Y chromosome has been refractory to the typical analytical approaches and remains almost completely unassembled and unexplored even when a fully finished reference assembly is available for the euchromatic genome. This is because the Y chromosome is entirely heterochromatic and densely packed with repetitive DNA, which confounds assembly. The euchromatic arms of anopheline chromosomes 2, 3 and X polytenize, but the heterochromatic Y does not, further complicating efforts to physically map and analyze this chromosome.
What little is currently known about the Y chromosome in Anopheles provides compelling reason to exhaustively identify its genes and other functional elements. First and foremost, the Y chromosome determines maleness, based on the fact that XXY carriers are male. Thus, it is presumed that the Y chromosome carries at least one factor important for sex determination. Identification of this factor(s) is a goal that would benefit both our understanding of the fundamental developmental genetics of anophelines, as well as vector control strategies where deliberate manipulation of gender is desirable. Second, the Y chromosome controls mating behavior, presumably due to additional factor(s) that remain to be discovered.
Interestingly, despite the paucity of protein coding genes, in Drosophila the Y chromosome has phenotypic effects on behavior, sex ratio, heat adaptation, and male fitness that may be epigenetic in origin. It is entirely unknown--but conceivable--that a similar situation might apply in Anopheles, and for this reason, understanding chromosome structure may be as important as exhaustively identifying transcribed genes on the Y.
We have recently formed a consortium between Virginia Tech and Notre Dame to improve our understanding of Y chromosome function, structure, and evolution in the An. gambiae complex.
Collaborators:
S. Emrich (Computer Science, Notre Dame), I. Sharakhov (Virginia Tech), J. Tu (Virginia Tech).