

Notre Dame Science
Hummon Research Group
Publications
2017
-
38)Lukowski JK, Weaver EM, Hummon AB. “Analyzing Liposomal Drug Delivery Systems in Three-Dimensional Cell Culture Models Using MALDI Imaging Mass Spectrometry”. Analytical Chemistry. 2017. In Press.
-
37)Boyce MW, LaBonia GJ, Hummon AB, Lockett MR. “Assessing chemotherapeutic effectiveness using a paper-based tumor model”. Analyst. 2017. In Press.
-
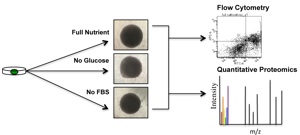
36)Schroll MM, LaBonia GJ, Ludwig KR, Hummon AB. “Glucose restriction combined with autophagy inhibition and chemotherapy in HCT 116 spheroids decreases cell clonogenicity and viability regulated by tumor supressor genes”. JPR. 2017. In Press.
-
35)Feist P, Sidoli S, Liu X, Schroll M, Rahmy S, Fujiwara R, Garcia B, Hummon AB. “Multicellular Tumor Spheroids Combined with Mass Spectrometric Histone Analysis to Evaluate Epigenetic Drugs” . Analytical Chemistry. 2017. Mar 7;89(5):2773-2781.
-
34)Ludwig KR, Hummon AB. “Mass Spectrometry for the Discovery of Biomarkers for Sepsis”. Molecular Biosystems. 2017. Mar 28;13(4):648-664.
2016
-
33)US Patent #9,464,328. “Biomarkers and Uses Thereof in Prognosis and Treatment Strategies for Right-Side Colon Cancer Disease and Left-Side Colon Cancer Disease” (Co-Inventor Steven Buechler).
-
32)Schunter A, Yue XS, Hummon AB. “Phosphoproteomics of Colon Cancer Metastasis: Comparative Mass Spectrometric Analysis of the Isogenic Primary and Metastatic Cell Lines SW480 and SW620”. Analytical and Bioanalytical Chemistry. 2016. Mar;409(7):1749-1763.
-
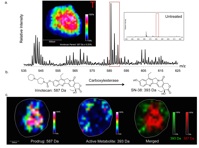
31)Liu X, Hummon AB."Chemical Imaging of Platinum-Based Drugs and their Metabolites". Scientific Reports. 2016. Dec 5;6:38507.
-
30)Yue XS, Lukowski JK, Weaver EM, Skube SB, Hummon AB. ”Quantitative Proteomic and Phosphoproteomic Comparison of 2D and 3D Colon Cancer Cell Culture Models”. Journal of Proteome Research. 2016. Dec 2;15(12):4265-4276..
-
29)Schroll M, Liu X, Herzog S, Skube SB, Hummon AB. “Nutrient Restriction of Glucose or Serum Results in Similar Proteomic Expression Changes in 3D Colon Cancer Cell Cultures” Nutrition Research. 2016. Oct;36(10):1068-1080.
28) Amanda B Hummon. “Toward Integrated Omics”. The Analytical Scientist, Issue 0616. 2016.
-
27)LaBonia GJ, Lockwood SY, Heller AA, Spence DM, Hummon AB. “Drug penetration and metabolism in 3-dimensional cell cultures treated in a 3D printed fluidic device: Assessment of irinotecan via MALDI imaging mass spectrometry”. PROTEOMICS. 2016. Jun;16(11-12):1814-21.
-
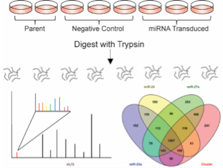
26)Ludwig KR, Dahl R, Hummon AB. “Evaluation of the mirn23a Cluster through an iTRAQ-based Quantitative Proteomic Approach”. Journal of Proteome Research. 2016. May 6; 15(5):1497-505.
2015
-
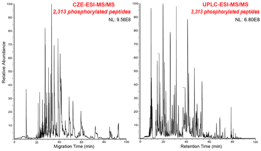
25)Ludwig KR, Sun L, Zhu G, Dovichi NJ, Hummon AB. “Over 2,300 Phosphorylated Peptide Identifications with Single-Shot Capillary Zone Electrophoresis-Tandem Mass Spectrometry in a 100 min Separation”. Analytical Chemistry. 2015. Sept 23; 87(19): 9532-9537.
-
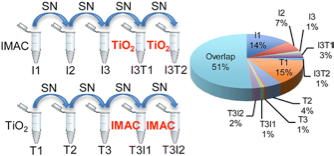
24)Yue XS, Schunter A, Hummon AB. “Comparing Multi-Step IMAC and Multi-Step TiO2 Methods for Phosphopeptide Enrichment”. Analytical Chemistry. 2015. August 3;87(17):8837-8844.
-
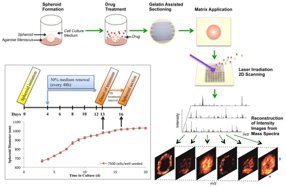
23)Liu X, Hummon AB. “Mass Spectrometry Imaging of Therapeutics from Animal Models to Three-Dimensional Cell Cultures”. Analytical Chemistry. 2015. Jun 18;87(19):9508-9519.
-
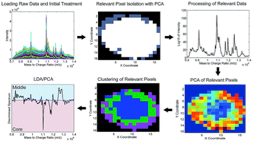
22)Weaver EM, Hummon AB, Keithley RB. “Chemometric Analysis of MALDI Mass Spectrometric Images of Three-Dimensional Cell Culture Systems”. Analytical Methods. 2015. Sept 7;7(17):7208-7219.
-
21)Feist P, and Hummon AB “Proteomic Challenges: Sample Preparation Techniques for Microgram-Quantity Protein Analysis from Biological Samples”. International Journal of Molecular Sciences. 2015. Feb 5;16(2):3537-63.
-
20)Feist P, Sun L, Liu X, Dovichi NJ, Hummon AB. "Bottom-up proteomic analysis of single HCT 116 colon carcinoma multicellular spheroids". Rapid Communications in Mass Spectrometry. 2015. Apr 15;29(7):654-658.
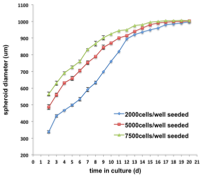
2014
-
19)Liu X, Hummon AB. "Quantitative Determination of Irinotecan and the Metabolite SN-38 by nLC-ESI-MS/MS in Different Regions of Multicellular Tumor Spheroids". Journal of The American Society for Spectrometry. 2014. Apr; 26(4):577-86.
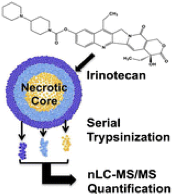
-
18)Bauer KM, Watts T, Buechler S, Hummon AB. "Proteomic and functional investigation of the colon cancer relapse-associated genes NOX4 and ITGA3". Journal of Proteome Research. 2014 Nov 7; 13 (11);4910-8.
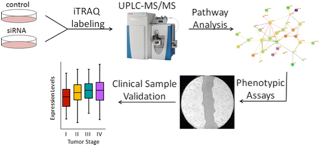
-
17)Ahlf DR, Masyuko RN, Hummon AB, Bohn PW. “Correlated Mass Spectrometry Imaging and Confocal Raman Microscopy for Studies of Three-Dimensional Cell Culture Sections”. Analyst. 2014 Sep 21;139 (18);4578-85.
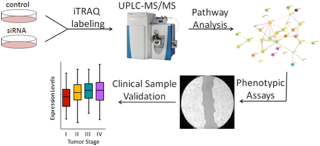
-
16)Wheatcraft Ahlf DR, Liu X, Hummon AB. “Sample Preparation Strategies for Mass Spectrometry Imaging of 3D Cell Culture Models”. Journal of Visualized Experiments. 2014 Dec 5;(94).
2013
-
15)Weston LA, Bauer KM, Skube SB, Hummon AB. “Selective, bead-based global peptide capture using a bifunctional crosslinker”. Anal Chem. 2013 Nov 19;85(22):10675-9.
-
14)Weston LA, Hummon AB. "Comparative LC-MS/MS analysis of Optimal Cutting Temperature (OCT) compound removal for the study of mammalian proteomes". Analyst. 2013 Nov 7;138(21):6380-4.
-
13)Keithley RB, Weaver EM, Metzinger MP, Rosado AM, Hummon AB, Dovichi NJ. “Single Cell Metabolic Profiling of Tumor Mimics.” Anal Chem. 2013 Oct 1;85(19);8910-8.
-
12)Yue XS, Hummon AB. "Combination of Multi-Step IMAC Enrichment with High-pH Reverse Phase Separation for In-Depth Phosphoproteomic Profiling". Journal of Proteome Research. 2013 Sep 6;12(9):4176-86.
-
11)Weston LA*, Bauer KM*, Hummon AB. "Comparison of bottom-up proteomic approaches for LC-MS analysis of complex proteomes". Analytical Methods, 2013, 5 (18), 4615 - 4621.
-
10)Liu, X, Weaver EM, Hummon AB."Evaluation of Therapeutics in Three-Dimensional Cell Culture Systems by MALDI Imaging Mass Spectrometry". Analytical Chemistry, 2013 Jul 2;85(13):6295-302. Epub 2013 Jun 11.
-
9)Hummon, A.B.; Dovichi, N.J. “The Tools of Proteomics” The Analytical Scientist, 2013, 1, 413-420.

-
8)Dovichi NJ, Hummon AB. "The Tools Behind Genomics" The Analytical Scientist, 2013, 1, 24-31.

-
7)Weaver EM, Hummon AB. "Imaging mass spectrometry: from tissue sections to cell cultures" Advanced Drug Discovery Reviews, 2013 Jul;65(8):1039-55.
2012
-
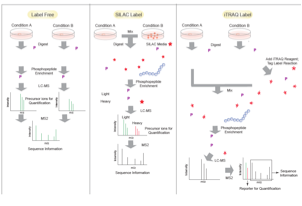
6)Yue XS, Hummon AB. "Mass spectrometry-based phosphoproteomics in cancer research." Front. Biol. 2012, 7(6): 566–586.
-
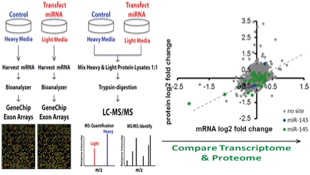
5)Bauer KM, Hummon AB. “Effects of the miR-143/-145 MicroRNA Cluster on the Colon Cancer Proteome and Transcriptome.” J Proteome Res. 2012 Sep 7;11(9):4744-54.
-
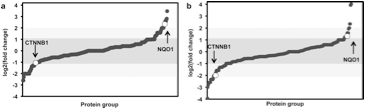
4)Bauer KM, Lambert PA, and Hummon AB. "Comparative label-free LC-MS/MS analysis of colorectal adenocarcinoma and metastatic cells treated with 5-fluorouracil" Proteomics 2012 Jun;12(12):1928-37.
-
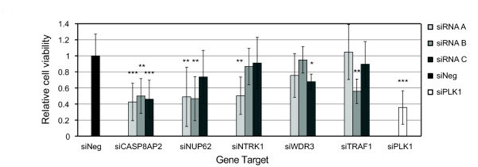
3)Hummon AB, Pitt JJ, Camps J, Emons G, Skube SB, Huppi K, Jones TL, Beissbarth T, Kramer F, Grade M, Difilippantonio MJ, Ried T and Caplen NJ. "Systems-wide RNAi analysis of CASP8AP2/FLASH shows transcriptional deregulation of the replication-dependent histone genes and extensive effects on the transcriptome of colorectal cancer cells." Molecular Cancer 2012 Jan 4;11:1.
-
2)Bauer, K.M.; Hummon, A.B.; Buechler, S. “Right-side and left-side colon cancer follow different pathways to relapse.” Mol Carcinog., 2012 May; 51(5);411-21.
2011
-
1)Li H, Hummon AB. “Imaging mass spectrometry of three-dimensional cell culture systems.” Anal Chem. 2011 Nov 15;83(22) 8794-801.
Post Doc and Graduate Training
-
18)Padilla-Nash HM, McNeil NE, Yi M, Nguyen QT, Hu Y, Wangsa D, Mack DL, Hummon AB, Case C, Cardin E, Stephens R, Difilippantonio MJ, Ried T. Aneuploidy, Oncogene Amplification, and Epithelial to Mesenchymal Transition Define Spontaneous Transformation of Murine Epithelial Cells."Carcinogenesis. 2013 Mar 15;73(6);2013-13.
-
17)Camps J, Pitt JJ, Emons G, Hummon AB, Case CM, Grade M, Jones TL, Nguyen QT, Ghadimi BM, Beissbarth T, Difilippantonio MJ, Caplen NJ, Ried T. ”Genetic Amplification of the NOTCH Modulator LNX2 Upregulates the WNT/β-Catenin Pathway in Colorectal Cancer.” Cancer Res. 2013 Mar 15;73(6):2003-13.
-
16)Grade, M.; Hummon, A.B.; Emons, G.; Camps, J.; Spitzner, M.; Hoermann, P.; Gaedcke, J.; Ebner, R.; Becker, H.; Ghadimi, B.M.; Beissbarth, T.; Difilippantonio, M.J.; Caplen, N.J.; Ried, T. “Identification and Validation of NOL5A and RPS2 as Potential Therapeutic Targets in Colorectal Cancer using a Functional Genomics Approach.” Int J. Cancer. March 2011 128(5):1069-79.
-
15)Camps, J.; Nguyen, Q.T.; Padilla-Nash, H.M.; Knutsen, T.; McNeil, N.E.; Wangsa, D.; Hummon, A.B.; Grade, M.; Ried, T.; Difilippantonio, M.J. “Integrative genomics reveals mechanisms of copy number alterations responsible for transcriptional deregulation in colorectal cancer.” Genes Chromosomes Cancer. 2009 Nov;48(11);1002-17.
-
14)Camps, J.; Grade, M.; Nguyen, Q.T.; Hörmann, P.; Becker, S.; Hummon, A.B.; Rodriguez, V.; Chandrasekharappa, S.; Chen, Y.; Difilippantonio, M.J.; Becker, H.; Ghadimi, B.M.; Ried, T. “Chromosome breakpoints in primary colon cancer cluster at sites of structural variants in the genome.” Cancer Res. 2008 Mar 1;68(5):1284-95.
-
13)Hummon, A.B.; Martin, S.E.; Caplen, N.J. “RNAi screens: Going right to the target.” Screening-Trends in Drug Discovery 4, 2 (2007).
-
12)Hummon, A.B.; Lim, S.R.; Difilippantonio, M.J.; Ried, T. “Isolation and solubilization of proteins after TRIzol extraction of RNA and DNA from patient material following prolonged storage.” Biotechniques 42, 467 (2007).
-
11)Grade, M.; Hörmann, P.; Becker, S.; Hummon, A.B.; Wangsa, D.; Varma, S.; Simon, R.; Liersch, T.; Becker, H.; Difilippantonio, M.J.; Ghadimi, B.M.; Ried, T. “Gene Expression Profiling Reveals a Massive, Aneuploidy-Dependent Transcriptional Deregulation and Distinct Differences between Lymph Node Negative and Positive Colon Carcinomas.” Cancer Res. 67, 41 (2007).
-
10)Hummon, A.B.; Richmond, T.A.; Amare, A.; Ewing, M.A.; Verleyen, P.; Baggerman, G.; Schoofs, L.; Southey, B.; Rodriguez-Zas, S.L.; Robinson, G.E.; Sweedler, J.V. “From the Genome to the Proteome: Uncovering Peptides in the Apis Brain.” Science 314, 647 (2006).
-
9)The Honeybee Genome Sequencing Consortium. "Insights into social insects from the genome of the honeybee Apis mellifera." Nature 443, 931 (2006).
-
8)Amare, A.; Hummon, A.B.; Southey, B.R.; Zimmerman, T.A.; Rodriguez-Zas, S.L.; Sweedler, J.V. “Bridging Neuropeptidomics and Genomics with Bioinformatics: Prediction of Mammalian Neuropeptide Prohormone Processing.” J. Proteome Res. 5, 1162 (2006).
-
7)Hummon, A.B.; Amare, A.; Sweedler, J.V. “Discovering New Invertebrate Neuropeptides using Mass Spectrometry.” Mass Spectrom. Rev. 25, 77 (2006).
-
6)Stuart, J.N.; Hummon, A.B.; Sweedler, J.V. “The Chemistry of Thought: Neurotransmitters in the Brain.” Anal. Chem. 76, 121A (2004).
-
5)Cummins, S.F.; Nichols, A.; Amare, A.; Hummon, A.B.; Sweedler, J.V.; Nagle, G.T. “Novel Candidate Protein Pheromones in Aplysia: Identification, Characterization, Expression, and Biological Activity.” J. Biol. Chem. 279, 25614 (2004).
-
4)Page, J.S.; Hummon, A.B.; Sweedler, J.V. “Single Cell Peptide Analysis with MALDI-MS” for Encyclopedia of Mass Spectrometry. Vol 2, Biological Applications. Eds. R. Caprioli and M. Gross. 2004.
-
3)Hummon, A.B.; Hummon, N.P.; Corbin, R.W.; Li, L.; Vilim, F.S.; Weiss, K.R.; Sweedler, J.V. “From Precursor to Final Peptides: A Statistical Sequence-Based Approach to Predicting Prohormone Processing.” J. Proteome Res. 2, 650 (2003).
-
2)Hummon, A.B.; Corbin, R.W.; Sweedler, J.V. “Discovering New Neuropeptides using Single Cell Mass Spectrometry.” Trends Anal. Chem. 22, 515 (2003).
-
1)Hummon, A.B.; Huang, H.; Kelley, W.P.; Sweedler. J.V. “A Novel Prohormone Processing Site in Aplysia californica: the Leu-Leu Rule.” J. Neurochem. 82, 1398 (2002).

