

Notre Dame Science
Hummon Research Group
Research
The Hummon research group seamlessly combines analytical chemistry and the study of cancer biology to explore gene products deregulated in the development and progression of cancer. Cancer arises from insults to the genome. With genomic damage, the expression levels of genes are altered from their normal state. Changes in the genome, transcriptome and proteome are highly conserved among samples from adenomas to carcinomas to metastases. Because genetic changes are commonly repeated among cancer patients, a better understanding of which genes, transcripts, and proteins are affected could have broad health implications. Therefore, the best way to understand the molecular underpinnings of cancer is to dissect the deregulated pathways that are contributing to the cancer phenotype, identify the aberrantly expressed genes and their products, and decipher their effect on downstream targets. The Hummon Research Group develops methods to evaluate both the transcriptome and the proteome in cancer cells.
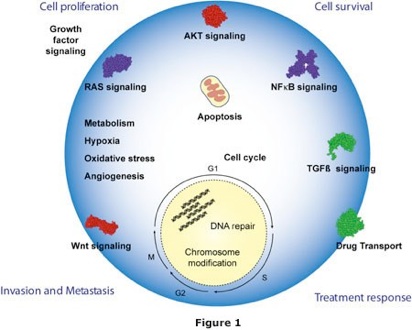
We develop and adapt current mass spectrometric and sampling protocols for global molecular profiling to understand cancer systems. Cancer cells are small, complex entities, differing from normal cells in their molecular equilibria (Figure 1). We need multidimensional analytical strategies to probe these cells to understand how and why they behave differently from normal cells. In order to accurately compare mRNA and protein levels, we need to generate high quality quantitative data.
Some of the current projects in the laboratory:
• Investigating the impact that individual and clusters of miRNAs have on the cancer proteome and transcriptome
• Examining changes to the phosphoproteome that accompany cancer progression
• Interrogating the distribution of analytes in three-dimensional cell cultures using imaging mass spectrometry
• Exploring changes to the proteome following RNA interference-based reduction of the expression of pivotal regulatory genes that have been shown to regulate the viability of colorectal cancer cells such as NOX4, ITGA3, and FLASH
•Developing novel sample preparation strategies for proteomic analysis
•Investigating the impact that individual and clusters of miRNAs have on the cancer proteome and transcriptome.
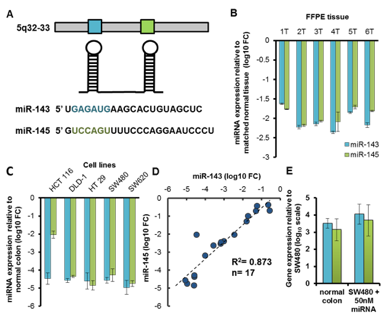
We investigated the effect that the miR-143/145 cluster has on the transcriptome and proteome of colorectal cancer. Expression of the miR-143/-145 cluster is reduced in colon cancer (Figure 3). Both miR-143 and miR-145 have been shown to possess antitumorigenic activity and are implicated in various cancer-related processes such as proliferation, invasion and migration. As the deregulation of the miR-143/145 cluster is implicated in tumorigenesis, we combined SILAC and microarray analyses to systematically interrogate the impact of miR-143/145 on the colon cancer proteome and transcriptome. Using SILAC we identified over 2000 proteins after reintroduction of miR-143 and miR-145, in the colon cancer cell line SW480, individually and then, in concert. Our goal was to determine whether these microRNAs function individually or synergistically. The resulting regulated gene products showed evidence of both mRNA destabilization and translational inhibition with a bias towards the former mechanism of regulation. Numerous candidate targets were identified whose expression is attributable to an individual microRNA or whose regulation was more apparent following reintroduction of the miR-143/145 cluster. In addition, several shared targets of miR-143 and miR-145 were identified. Overall, our results indicate that the summed effects of individually introduced microRNAs produce distinct molecular changes from the consequences of the assembled cluster. This finding has broad implications to additional clusters in a number of disease states and cellular processes. We conclude that there is a need to investigate both the individual and combined functional implications of a microRNA cluster.
Figure 3. miR-143 and miR-145 expression levels in colon cancer. (A) The miR-143-145 cluster located on Chr 5q32-33. (B) qRT-PCR results for miR-143 and miR-145 expression level in six primary formalin-fixed, paraffin-embedded (FFPE) normal colon and patient matched colon tumor tissue samples (C) qRT-PCR results for miR-143 and miR-145 expression levels in five human colon cancer cell lines. (D) Scatter plot of miR-145 versus miR-143 levels in cell lines and FFPE samples with linear regression line (R2= 0.873, P<0.0001, n=17). (E) qRT-PCR results for miR-143 and miR-145 expression levels in normal colon mucosa and SW480’s after overexpression of 50nM miR-143 or miR-145. Data represented as median ± S.D. (B, C, E).
For more information about this study, please see: Bauer KM, Hummon AB. “Effects of the miR-143/-145 MicroRNA Cluster on the Colon Cancer Proteome and Transcriptome.” J Proteome Res. 2012;11(9):4744-54.
•Examining changes to the phosphoproteome that accompany cancer progression
We are investigating alterations in the phosphoproteome that occur at two critical points in cancer progression. First, we are examining the changes in the phosphoproteome that occur with exposure to very low doses of ionizing radiation, which can lead to tumorigenesis. We are also comparing changes in the abundance of phosphoproteins in primary versus metastatic colon cancer cells. Ionizing radiation (IR) is a well-known cause for DNA damage. IR provides energy to the DNA backbone resulting in double strand breakage. Many of the DNA damage response pathways are activated through phosphorylation or dephosphorylation after DNA damage, either to repair the damaged DNA backbone or to initiate cell death pathways. Previous studies have reported that high doses of IR exposure cause cell death. We hypothesize that under much lower doses of IR exposure, the damage may be prone to initiate pathways to repair DNA damage. We are exposing immortalized, non-tumorigenic cells to very low doses of IR and then evaluating the changes to the phosphoproteome. After IR exposure, phosphopeptides are selectively enriched with strong cation exchange (SCX) chromatography followed by immobilized metal affinity (IMAC) beads, and then analyzed by mass spectrometry (Figure 4). By monitoring the changes in the phosphoproteome at different time points after IR, the process of DNA damage repair is analyzed and the key factors contributing to cancer development can be determined. This study will provide useful information for IR safety and cancer treatment.
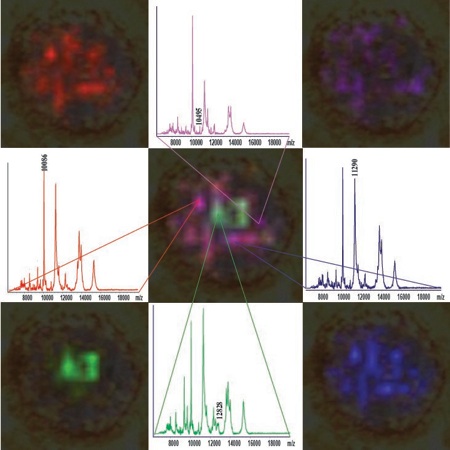
Figure 5. Mass spectra and m/z ion maps for analytes detected in a section of a 3D colon culture, also known as a spheroid.
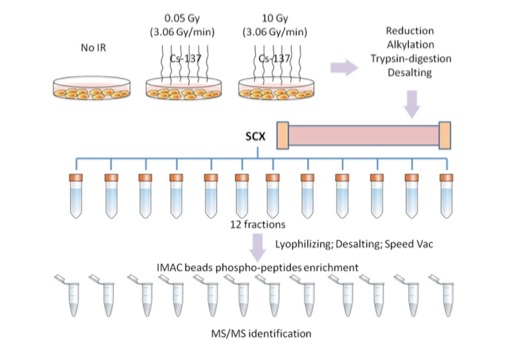
Figure 4. Phosphoproteomic workflow to identify proteins altered by ionizing radiation.
Using the same workflow, we are also investigating the quantitative changes in the phosphoproteome in primary and metastatic colon cancer cells to identify pathways that are altered in metastatic progression. A patient-matched set of Stage II and Stage III colon cancer cell lines are labeled with stable isotopes (SILAC), enriched for phosphopeptides, and then analyzed by LC-MS/MS to examine the molecular differences between primary and metastatic colon cancer.
More information about phosphoproteomic analysis by mass spectrometry can be found in our recent paper: Yue XS, Hummon AB. "Mass spectrometry-based phosphoproteomics in cancer research." Front. Biol. 2012, 7(6): 566–586.
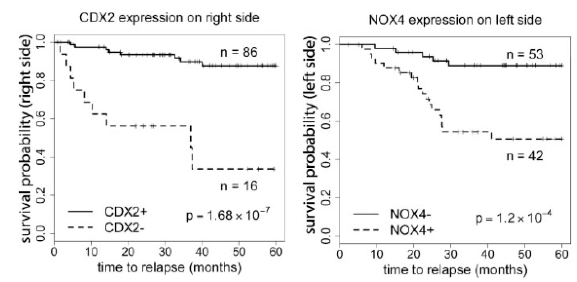
Figure 6. Survival plots for right and left sided colon cancer using CDX2 and NOX4 expression in primary colon cancer. Plots are based on previously published microarray data.
-
•Interrogating the distribution of analytes in three-dimensional (3D) cell cultures using imaging mass spectrometry
Three dimensional cell cultures are popular model systems in health-related biological research. They combine the flexibility of cell culture with structural information not possible with standard two-dimensional cultures. Imaging analytes in three-dimensional cell culture systems is typically accomplished via microscopy or immunological approaches. With these classic approaches, the analytes are preselected before analysis, limiting discovery-based applications. Our research group is developing tools to enable imaging mass spectrometry analysis for the characterization of three-dimensional cell culture systems. Mass spectrometric profiling does not require preselection of analytes.
We are the first research group to examine three dimensional cell culture systems via imaging mass spectrometry. We are developing approaches to manipulate 3D cultures and prepare them for imaging. We are working with colon adenocarcinoma cell lines, which form 3D structures called spheroids. In our mass spectrometric images, we detect changes in the spatial distribution of proteins throughout the spheroid structures (Figure 5). We will expand our studies to genetically manipulated 3D cultures and explore molecular changes associated with the beginning of metastasis and the epithelial to mesenchymal transition. To examine the changes in protein expression and distribution that accompany this transition, we are growing cultures containing inducible shRNAs that silence a critical regulatory gene, E-Cadherin. We will map the alterations in protein expression that accompany silencing of E-Cadherin on the protein level. Finally, we are applying our approach to examine drug concentrations and penetration depth into 3D cultures.
For more information on our imaging work, please see Li H, Hummon AB. “Imaging mass spectrometry of three-dimensional cell culture systems.” Anal Chem. 2011 Nov 15;83(22) 8794-801 and Weaver EM, Hummon AB. "Imaging mass spectrometry: from tissue sections to cell cultures" Advanced Drug Discovery Reviews. 2013 In Press.
-
•Exploring changes to the proteome following RNA interference-based reduction of the expression of pivotal regulatory genes that have been implicated in the progression of colorectal cancer such as NOX4 and FLASH
We are using loss-of-function analysis via RNA interference to investigate the role of specific genes in colorectal cancer. For example, we are examining the changes to the transcriptome and proteome following the reduction of the gene FLASH. Knockdown of FLASH has been shown to drastically reduce the viability of colon cancer cells.
We have also investigated the distinct gene expression patterns on the right and left side of the colon and their relationship to relapse. In particular, expression of the genes CDX2 and NOX4 are strong predictors of survival (Figure 6). We are further exploring the functional role of NOX4 with in vitro loss-of-function studies.
•Developing novel sample preparation strategies for proteomic analysis
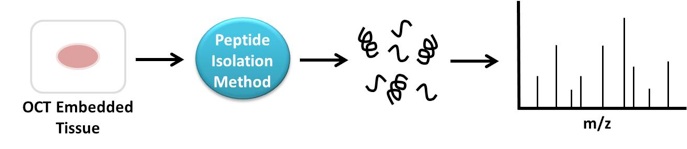
As a complement to our mechanistic studies, we are also developing proteomic sample preparation protocols. We have devised an improved method for phosphopeptide enrichment. We are also developing a novel method to selectively enrich and isolate peptides from archived tissue samples embedded in optimal cutting temperature (OCT) compound (Figure 7). Many primary bio-banked tissue samples are embedded in OCT, a substance that interferes with mass spectrometric analysis. Successful analysis of these bio-banked tissue samples would be an important resource in understanding cancer.
For more information on our studies of CDX2 and NOX4, please see Bauer, K.M.; Hummon, A.B.; Buechler, S. “Right-side and left-side colon cancer follow different pathways to relapse.” Mol Carcinog., 2012, 51(5);411-21.
Figure 7. Schematic of peptide enrichment approach to isolate peptides
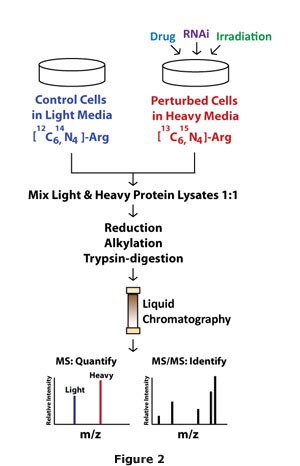
While advancing methodologies to simultaneously probe global mRNA and protein levels, we also incorporate a loss of function approach to identify the role of individual genes contributing to the cancer phenotype. Even with the high level of annotation of the human genome, the functional role of most genes remains unknown. We use RNA interference and other controlled perturbations to examine gene function in cancer (Figure 2).
Individual genes are perturbed, either with RNAi-based approaches or small molecule inhibitors, and then mRNA and protein profiles are generated. To profile the cancer transcriptome and proteome, we utilize gene expression microarray and quantitative Stable Isotope Labeling by Amino acids in Cell culture (SILAC) mass spectrometry protocols in cell lines. Measurement and determination of the mRNA and protein profiles expose pivotal imbalances and downstream gene targets in cancer, revealing windows for potential therapeutic manipulation.

