| Severson Lab Home |
| Biological Sciences Home |

|
 |
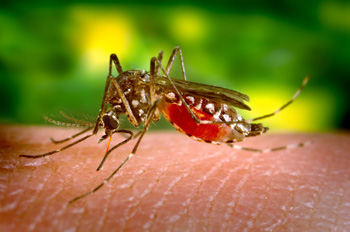
The Aedes aegypti Genome Project |  |
We conducted an NIH NIAID funded project (UO1 AI050936) that has produced a complete annotated genome sequence for the yellow fever mosquito Aedes aegypti. Our initial intent for this project was to develop a body of genome information including EST sequences, genomic sequences, and the physical map positions of large genomic clones that will enhance gene discovery, and also provide critical front-end tools for the whole-genome sequencing effort. My collaborators were Dr. Dennis Knudson at Colorado State University, Drs. Brendan Lofus and Vish Nene at The Institute for Genome Research (TIGR), and Dr. Bento Soares at the University of Iowa.
Details on the genome assembly and annotation were published in the  June 22, 2007 issue of Science (316:1718-1722).
June 22, 2007 issue of Science (316:1718-1722).
Our Plans for the Aedes aegypti genome project are described in the Project Plan.
The annotated Aedes aegypti genome sequence is displayed for public access and query at VectorBase.
Please feel free to contact me for further information.