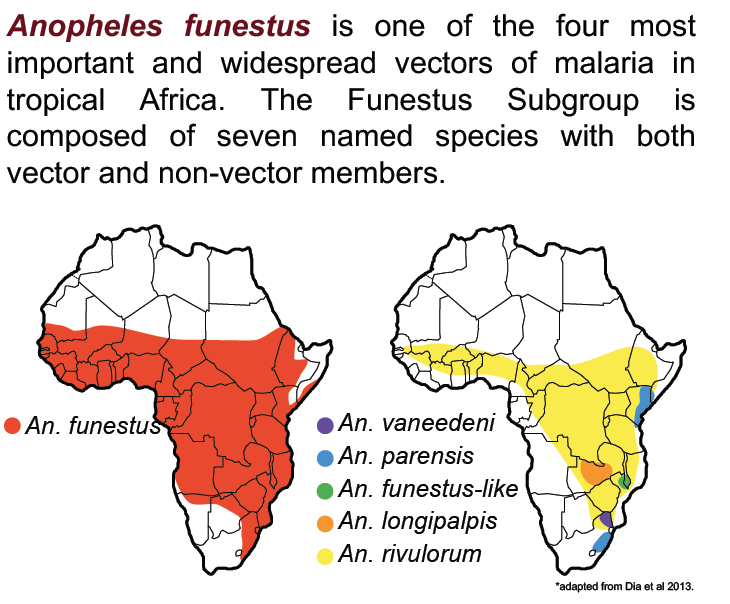
Elucidating the An. funestus species complex
Anopheles funestus is one of the most important and widespread vectors of human malaria in tropical Africa, but unlike An. gambiae, it is understudied. This major vector conceals a group of close relatives that are morphologically similar or identical as adults. None of these species, with rare exceptions, has been directly implicated as a secondary vector. However, they can rest indoors in large numbers and are considered important due to their impact on scarce resources available for vector control. They overlap with An. funestus, but their geographic distribution is largely unknown, and their actual or potential contribution to malaria transmission is also uncertain. Traditionally characterized as zoophilic and exophilic, their behavior is plastic and depends upon host availability and other (unknown) environmental factors. Moreover, the taxonomic complexity of this group is at least as high as the better-known An. gambiae complex; within each named (or unnamed) species are genetically heterogeneous clusters whose interrelationships have never been resolved, and additional cryptic diversity is being reported by vector control programs.
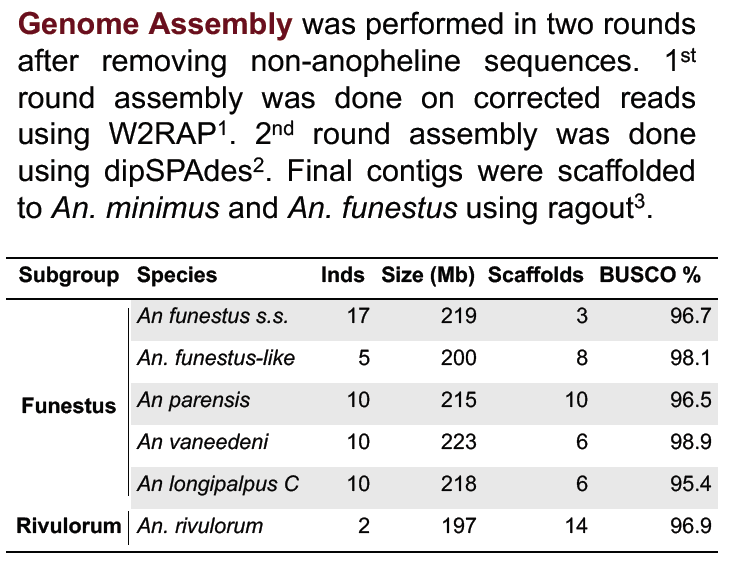
Our central goal in this project is to elucidate the An. funestus complex. Toward this end, we have generated draft reference genome assemblies for five species. Using whole genome alignments and additional light whole genome sequencing, we are constructing an initial portrait of species relationships. Similar to the An. gambiae complex, our data reveal multiple historical introgression events between different species pairs, suggesting that hybridization and introgression may be common during the radiation of anopheline species complexes. We have also stumbled upon what appears to be an undescribed new species in the An. funestus complex. Overall, these data will provide an essential foundation for unraveling the complex taxonomy, systematics and ultimately the biology of this important but poorly understood group.Funding: NIH NIAID R21 AI123491
Collaborators:
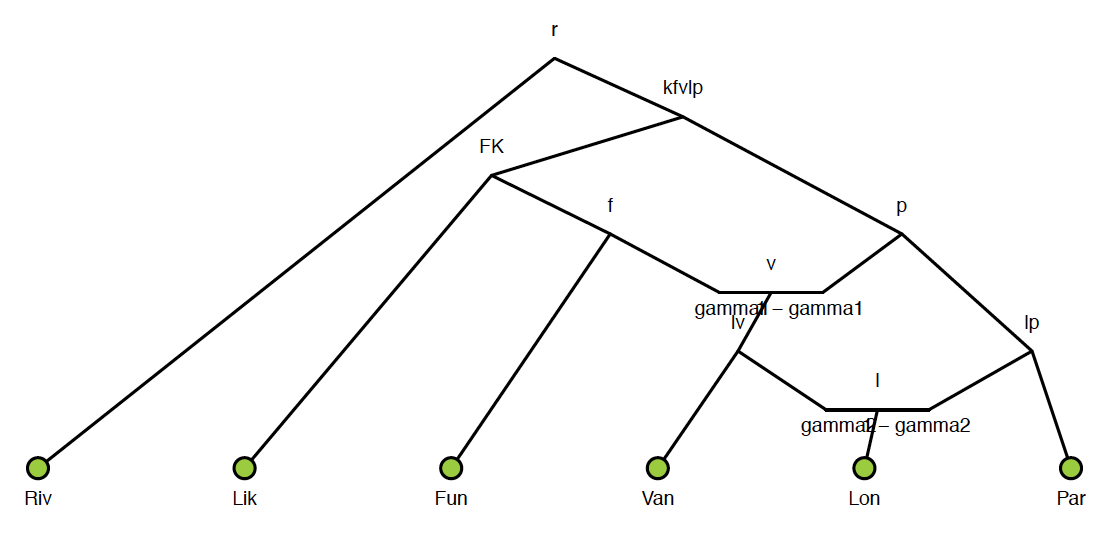 Admixture graph. A model of the best fit evolutionary history for the An funestus subgroup. Connecting lines and nodes represent admixture events between subgroup species.
Admixture graph. A model of the best fit evolutionary history for the An funestus subgroup. Connecting lines and nodes represent admixture events between subgroup species.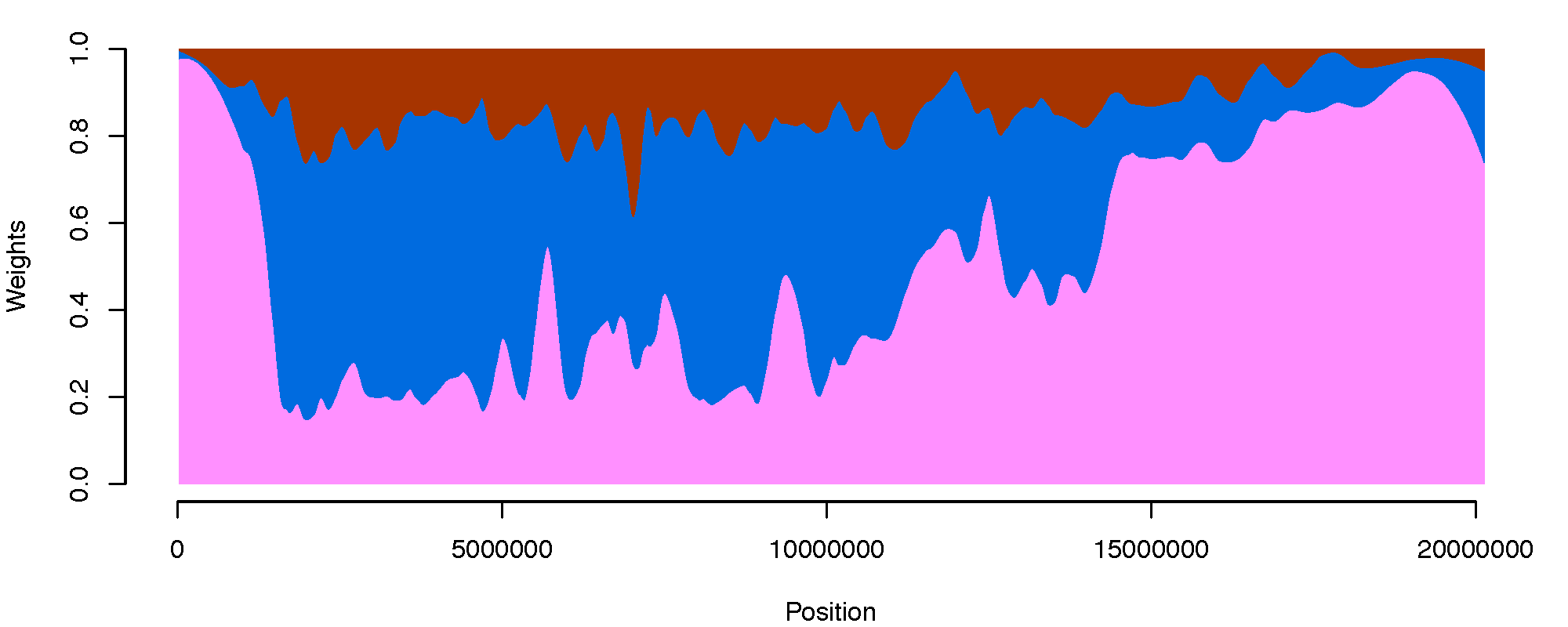 Chromosome painting. Distribution of gene tree phylogenies across the X chromosome reconstructed using 6 species of the An. funestus subgroup. Areas of single color represent a common gene topology. Regions composed of multiple colors represent discordant gene topologies with frequencies equal to colored area. Regions with discordant gene topologies are indicative of either incomplete lineage sorting or introgression between subgroup members.
Chromosome painting. Distribution of gene tree phylogenies across the X chromosome reconstructed using 6 species of the An. funestus subgroup. Areas of single color represent a common gene topology. Regions composed of multiple colors represent discordant gene topologies with frequencies equal to colored area. Regions with discordant gene topologies are indicative of either incomplete lineage sorting or introgression between subgroup members.