Chromosomal Inversion Polymorphisms in Anopheline Malaria Vectors
Molecular and Computational Karyotyping: Foundational genomic resources for novel vector-based strategies exist for An. gambiae and An. funestus (Vectorbase; Ag1000G/MalariaGen). Together, they enable detailed population genomics and genome-wide association studies (GWAS) aimed at understanding the genetic basis of epidemiologically important traits. However, these vector species are highly polymorphic for chromosomal inversions. Failing to account for inversions can mislead population genetic and genome-wide association studies and obscure relationships between inversions and epidemiologically relevant traits. Yet despite rapid advances in genome technology, classical cytogenetic determination of inversion status is the only method currently available. Inversion status is not obvious from genome re-sequencing data, as alleles are mapped to their position on a reference genome and not to their actual physical locations. Unfortunately, cytogenetic analysis is impractical or even prohibitive.
Addressing this gap, one of our central goals is to develop and validate computational and molecular inversion genotyping approaches, enabling a modern assessment of the association between inversions and epidemiologically relevant traits. We are approaching this goal through complementary strategies. Our main approach is whole genome sequencing of cytogenetically karyotyped An. gambiae and An. funestus, followed by computational SNP analysis to identify “tag SNP markers” inside the inverted region, whose allelic states are predictive of karyotype status. Both computational and molecular genotyping strategies are then developed from these tag SNPs. In collaboration with Russ Corbett, we are also applying innovative genomic technologies to molecularly characterize previously unknown inversion breakpoints, information that can be exploited in the design of molecular assays specifically targeting these breakpoints.
Functional Genomics and Systems Biology of Inversions: Inversion polymorphisms are responsible for many ecologically important phenotypes, and are often found under balancing selection. Indeed, they play an important role in local adaptation across taxonomically diverse species. In An. gambiae, two polymorphic inversions on chromosome 2 (denoted 2La and 2Rb) are associated with arid and hot conditions in Africa. They are maintained in spatially and temporally heterogeneous environments, and segregate along climatic gradients of aridity. Despite indirect evidence of their adaptive significance, little is known of the phenotypic targets of selection or the underlying genetic mechanisms. However, the same features that ensure their large role in local adaptation—especially reduced recombination between alternate arrangements—mean that uncovering the precise loci within inversions that control these phenotypes is unachievable using standard mapping approaches.
Accordingly, we have adopted a variety of alternative approaches to identify causal loci. These include (i) population pool sequencing from the endpoints and center of inversion clines; (ii) pool-GWAS; and (iii) systems genetics.
From population pool sequencing, we found significantly elevated differentiation between populations at opposite ends of the cline only in rearranged (not collinear) genomic regions, consistent with spatially varying selection maintaining the inversion cline in the face of unobstructed population connectivity. Importantly, nucleotide polymorphisms within the inversions that are fixed or strongly skewed for alternative alleles between endpoint populations showed patterns suggesting large amounts of gene flux between inverted and standard arrangements at the center of the cline, where the inversions are at intermediate frequencies and inversion heterozygotes are abundant.
In the pool-GWAS approach, we took advantage of long-term balancing selection on a pair of inversions to map desiccation tolerance. After measuring thousands of wild-caught individuals for survival under desiccation stress, we used phenotypically extreme individuals carrying alternative arrangements at the 2La inversion to construct pools for whole-genome sequencing. Genome-wide association mapping using these pools revealed dozens of significant SNPs within both 2La and 2Rb, many of which neighbored genes controlling ion channels or related functions.
In the systems genetics approach, we analysed the same two inversions. We profiled physiology, behaviour, and transcription in four different karyotypic backgrounds derived from a common parental colony. Acclimation to different climatic regimes resulted in pervasive inversion-driven phenotypic differences, whose magnitude and direction depended upon gender, environment, and epistatic interactions between inversions. Inversion-affected loci were significantly enriched inside inversions, as predicted by local adaptation theory. Drug perturbation supported lipid homeostasis and energy balance as inversion-regulated functions, a finding supported by research on climatic adaptation in multiple systems.
Our results point to the promise of similar approaches in systems with inversions maintained by balancing selection, and taken together, provide a list of candidate genes underlying the specific phenotypes controlled by the two inversions studied here. We are functionally validating those candidate genes using CRISPR and RNAi.
Funding: NIH R01 AI125360
References:
Collaborators (Past and Present):
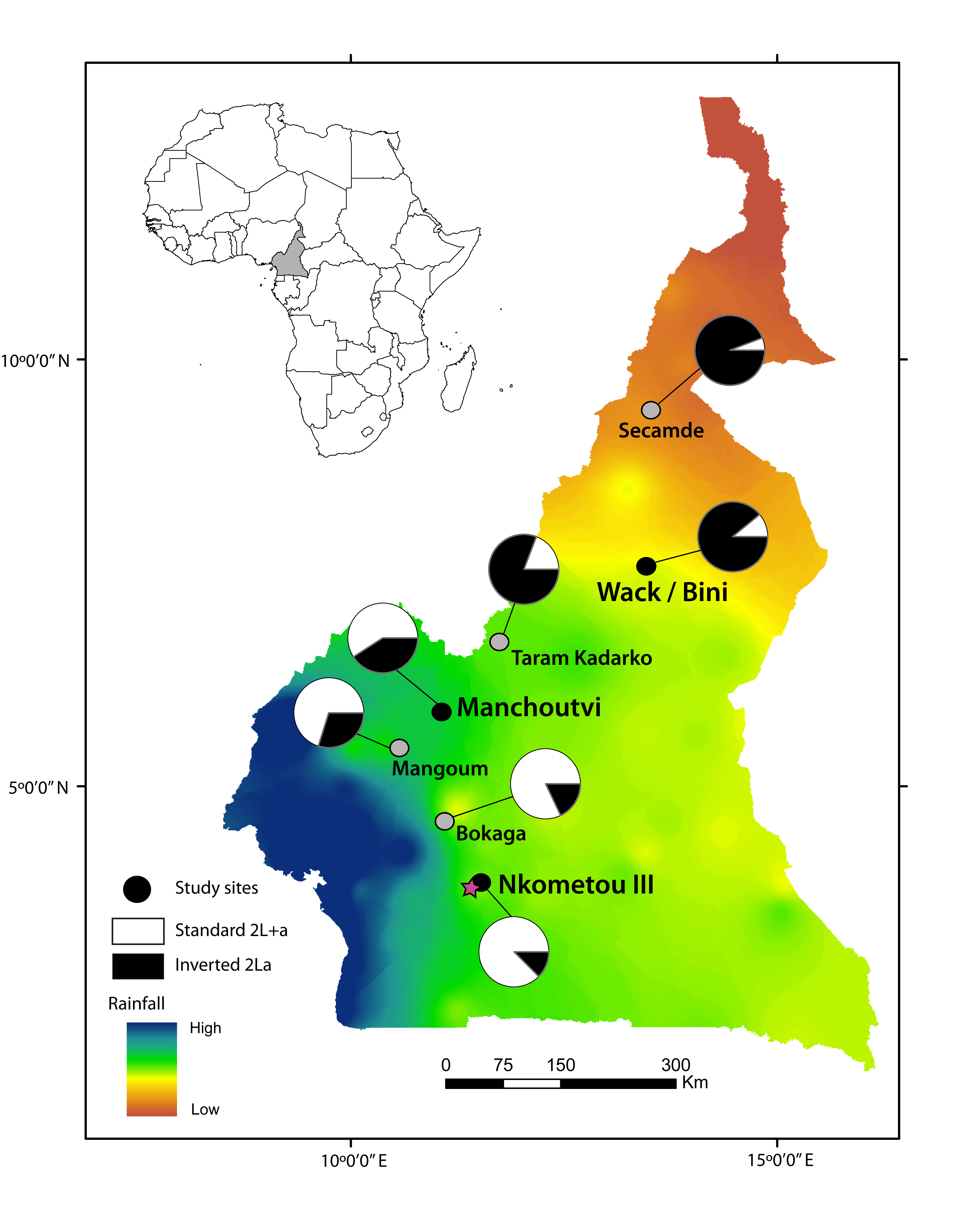 Fig. 1. Sampling sites along a latitudinal transect in Cameroon.
Fig. 1. Sampling sites along a latitudinal transect in Cameroon.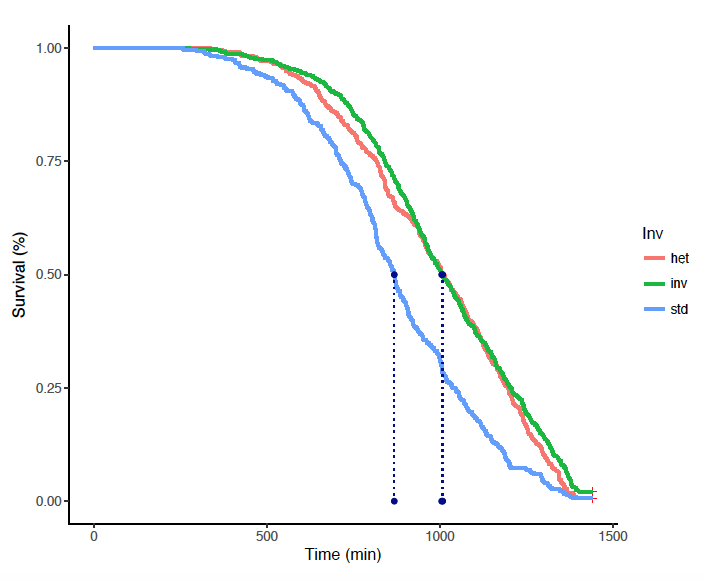 Fig. 2. Survival of teneral adult female An. gambiae under acute desiccation stress. Data are stratified by In(2La) karyotype.
Fig. 2. Survival of teneral adult female An. gambiae under acute desiccation stress. Data are stratified by In(2La) karyotype.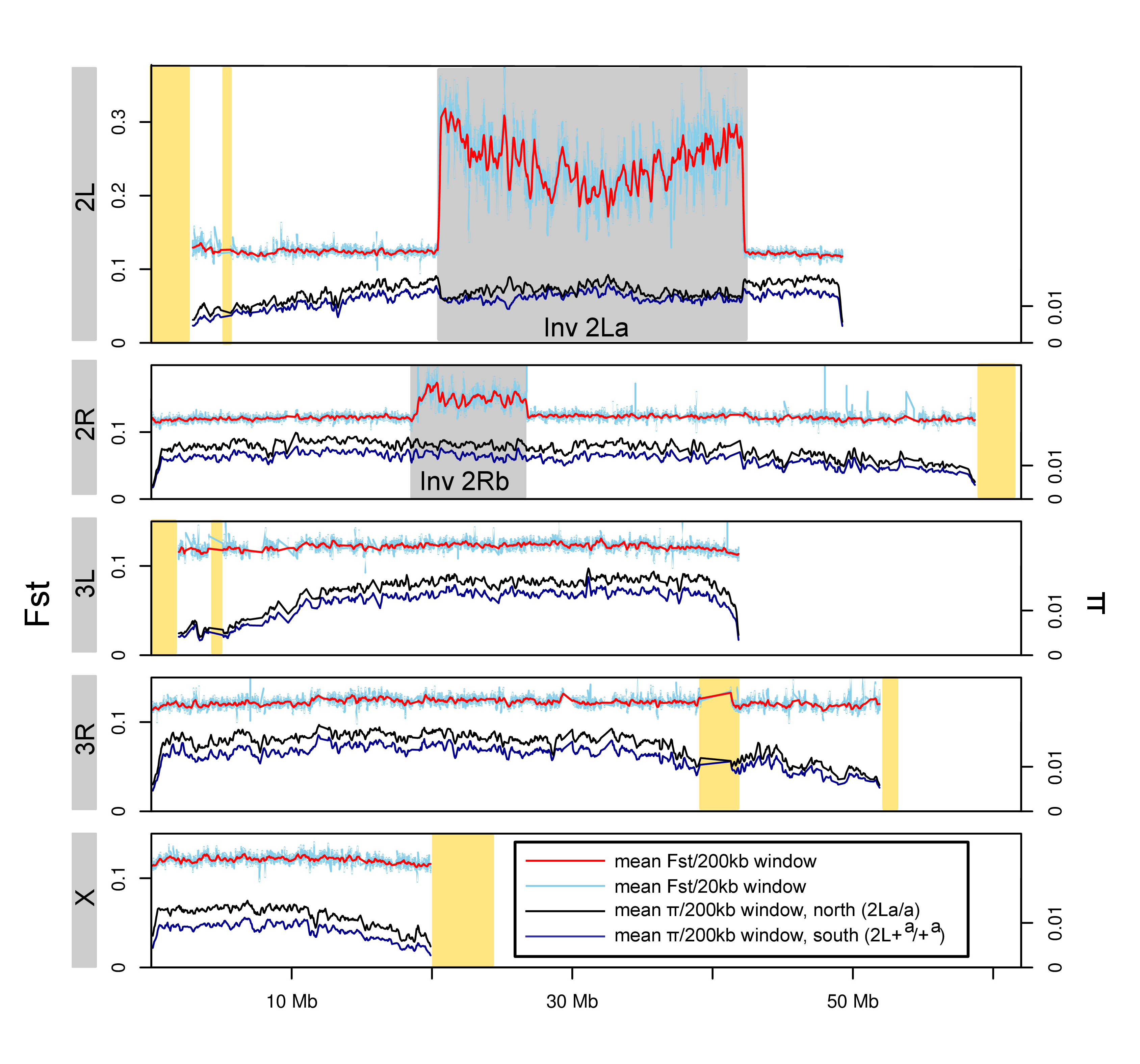 Fig. 3. Divergence (FST) between and diversity (p) within An. gambiae S populations from opposite ends of the latitudinal cline in Cameroon.
Fig. 3. Divergence (FST) between and diversity (p) within An. gambiae S populations from opposite ends of the latitudinal cline in Cameroon.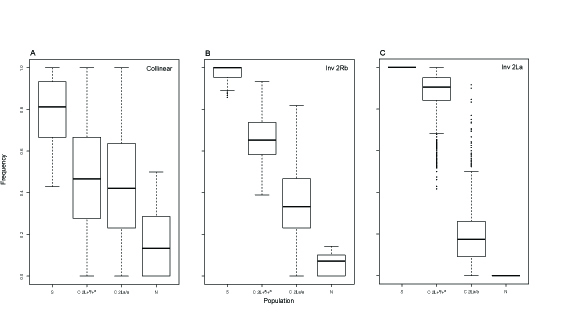 Fig. 4. Individual SNP frequencies along the latitudinal gradient in Cameroon, based on loci that most strongly differentiated populations at opposite ends of the gradient.
/>
Fig. 4. Individual SNP frequencies along the latitudinal gradient in Cameroon, based on loci that most strongly differentiated populations at opposite ends of the gradient.
/>
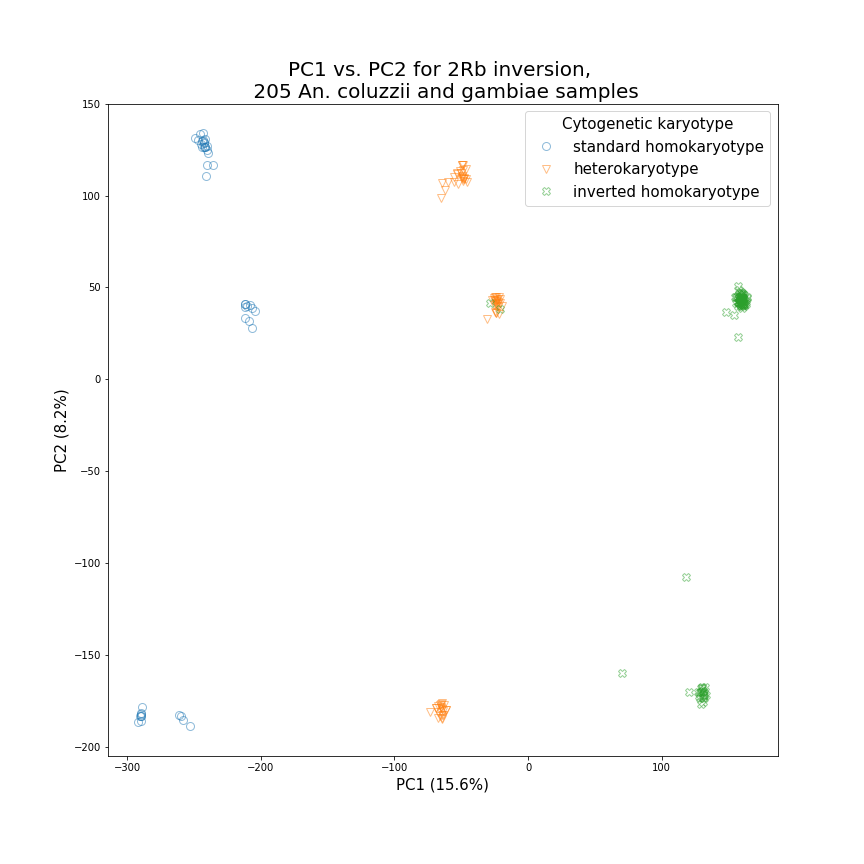 Fig. 5. Principal components analysis (PCA) recapitulates known patterns of chromosomal inversion karyotypes in An. coluzzii and An. gambiae. Using data from the 2Rb inversion, the first principal component among 205 An. coluzzii and An. gambiae from western and central Africa corresponds to cytogenetically obtained karyotypes. The second principal component corresponds to inter- and intra-species differentiation.
Fig. 5. Principal components analysis (PCA) recapitulates known patterns of chromosomal inversion karyotypes in An. coluzzii and An. gambiae. Using data from the 2Rb inversion, the first principal component among 205 An. coluzzii and An. gambiae from western and central Africa corresponds to cytogenetically obtained karyotypes. The second principal component corresponds to inter- and intra-species differentiation.
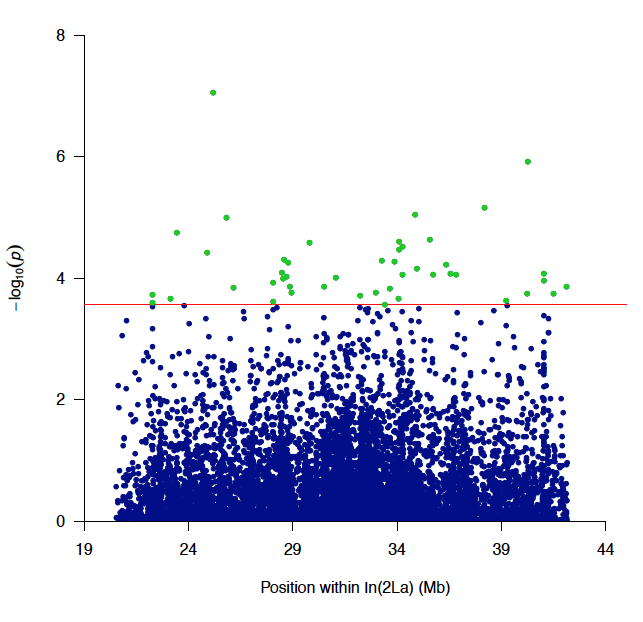 Fig. 6. Manhattan plot of the association P-values for acute desiccation tolerance inside In(2La). Horizontal red line represents the In(2La)-wide significance threshold at FDR of 5%. Tested SNPs below that threshold shown as blue dots; green dots represent significant SNPs conferring high tolerance.
Fig. 6. Manhattan plot of the association P-values for acute desiccation tolerance inside In(2La). Horizontal red line represents the In(2La)-wide significance threshold at FDR of 5%. Tested SNPs below that threshold shown as blue dots; green dots represent significant SNPs conferring high tolerance.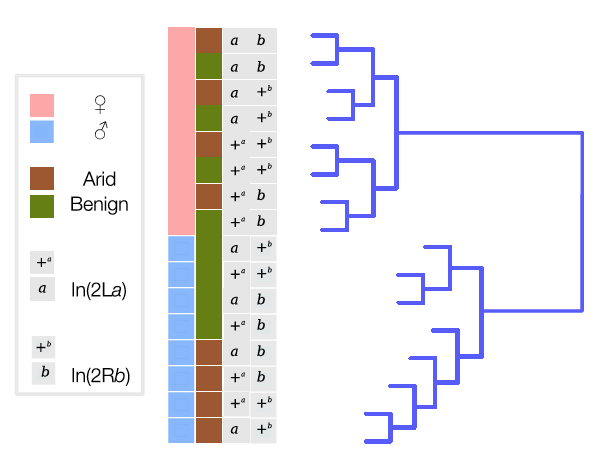 Fig. 7. Hierarchical clustering dendrogram of gene expression data. Terminal branches (samples) are labeled by gender (pink, female; blue, male), environmental condition (green, benign; brown, arid), and inversion status (+, standard; a or b, inverted).
Fig. 7. Hierarchical clustering dendrogram of gene expression data. Terminal branches (samples) are labeled by gender (pink, female; blue, male), environmental condition (green, benign; brown, arid), and inversion status (+, standard; a or b, inverted).