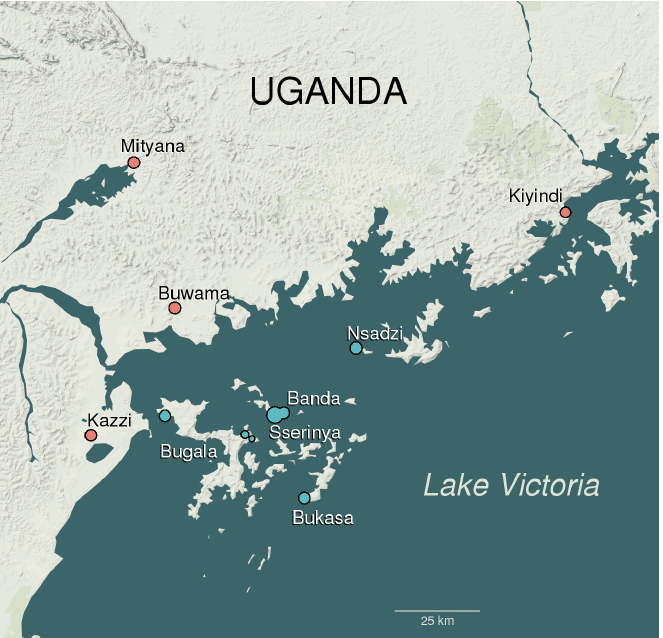
 Fig1. Map of study area showing sampling localities on Ssese Islands (blue) and mainland localities in Lake Victoria Basin (red). Map data copyright 2018 Google.
Fig1. Map of study area showing sampling localities on Ssese Islands (blue) and mainland localities in Lake Victoria Basin (red). Map data copyright 2018 Google.
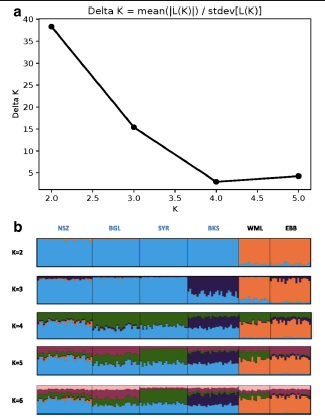 Fig2. Bayesian clustering analysis of combined 2002 microsatellite and mtDNA data from the same mosquito specimens. (a) The optimal number of clusters (K) was two based on the method of G Evanno, S Regnaut and J Goudet. (b) Bar plots with individual mosquitoes represented as vertical bars colored in proportion to their assignment to clusters inferred at each value of K (from 2 to 6). Individuals from the same population sample are grouped, with groups bounded by vertical black lines. Population labels are in blue font for islands and black font for mainland samples.
Fig2. Bayesian clustering analysis of combined 2002 microsatellite and mtDNA data from the same mosquito specimens. (a) The optimal number of clusters (K) was two based on the method of G Evanno, S Regnaut and J Goudet. (b) Bar plots with individual mosquitoes represented as vertical bars colored in proportion to their assignment to clusters inferred at each value of K (from 2 to 6). Individuals from the same population sample are grouped, with groups bounded by vertical black lines. Population labels are in blue font for islands and black font for mainland samples.
Population Biology of An. gambiae on Lake Victoria Islands
Understanding population genetic structure in the malaria vector An. gambiae is crucial to inform genetic control and manage insecticide resistance. Unfortunately, species characteristics such as high nucleotide diversity, large effective population size, recent range expansion, and high dispersal ability complicate the inference of genetic structure across its range in sub-Saharan Africa. The ocean, along with the Great Rift Valley, is one of the few recognized barriers to gene flow in this species, but the effect of inland lakes, which could be useful sites for initial testing of genetic control strategies, is relatively understudied. We are examining Lake Victoria as a barrier between the Ugandan mainland and the Ssese Islands, which lie up to 60 km offshore. We are applying indirect genetic estimates (including mtDNA, reduced-representation sequencing, and whole genomic sequencing) as well as direct ecological and entomological measures to better understand the island biology of An. gambiae in relation to disease transmission.
Funding:
Support for this work comes from Target Malaria, which receives core funding from the Bill & Melinda Gates Foundation and from the Open Philanthropy Project Fund, an advised fund of Silicon Valley Community Foundation.
References:
Bergey CM, Lukindu M, Wiltshire RM, Fontaine MC, Kayondo J, Besansky NJ (2018) Structure of selected variation in island Anopheles gambiae Giles (Diptera: Culicidae) and implications for genetic control field trials. In preparation.
Wiltshire RM, Bergey CM, Kayondo JK, Birungi J, Mukwaya LG, Emrich SJ, Besansky NJ, Collins FH (2018) Reduced-representation sequencing identifies small effective population sizes of Anopheles gambiae in the northwestern Lake Victoria basin, Uganda. Malaria J, in revision.
Lukindu M, Bergey CM, Wiltshire RM, Small ST, Bourke B, Kayondo JK, Besansky NJ (2018) Spatio-temporal genetic structure of Anopheles gambiae in the Northwestern Lake Victoria Basin, Uganda: implications for genetic control trials in malaria endemic regions. Parasites & Vectors 11: 246.
Collaborators (Past and Present):
University of Notre Dame: Martin Lukindu, Christina Bergey, Rachel Wiltshire, Scott Emrich, Frank Collins, Scott Small, Brian Bourke, Nora Besansky
Uganda Virus Research Institute: Josephine Birungi, Louis Mukwaya, Jonathan Kayondo
University of Groningen: Michael Fontaine
 Fig1. Map of study area showing sampling localities on Ssese Islands (blue) and mainland localities in Lake Victoria Basin (red). Map data copyright 2018 Google.
Fig1. Map of study area showing sampling localities on Ssese Islands (blue) and mainland localities in Lake Victoria Basin (red). Map data copyright 2018 Google.
 Fig2. Bayesian clustering analysis of combined 2002 microsatellite and mtDNA data from the same mosquito specimens. (a) The optimal number of clusters (K) was two based on the method of G Evanno, S Regnaut and J Goudet. (b) Bar plots with individual mosquitoes represented as vertical bars colored in proportion to their assignment to clusters inferred at each value of K (from 2 to 6). Individuals from the same population sample are grouped, with groups bounded by vertical black lines. Population labels are in blue font for islands and black font for mainland samples.
Fig2. Bayesian clustering analysis of combined 2002 microsatellite and mtDNA data from the same mosquito specimens. (a) The optimal number of clusters (K) was two based on the method of G Evanno, S Regnaut and J Goudet. (b) Bar plots with individual mosquitoes represented as vertical bars colored in proportion to their assignment to clusters inferred at each value of K (from 2 to 6). Individuals from the same population sample are grouped, with groups bounded by vertical black lines. Population labels are in blue font for islands and black font for mainland samples.