Genetic Underpinnings of Larval Saltwater Tolerance in the An. gambiae complex
In Family Culicidae, salinity tolerance in the larval stage has arisen repeatedly and recently in several lineages. Within genus Anopheles, malaria vector species in Africa, Europe, Southeast Asia, Australasia and the Neotropics are often embedded in species complexes whose members have contrasting larval ecologies, salinity tolerance, and roles in human malaria transmission. Clearly, salinity tolerance plays a key role in determining habitat use and ecological distribution of mosquitoes, and thus their contribution to malaria transmission.
There is an established and growing literature on the physiology of osmotic and ionic regulation in individual mosquito species, especially culicines. Yet surprisingly little is known regarding the genetic basis of this trait in mosquitoes. The ability to dissect the genetic basis of this adaptive trait using next generation genomic tools lays the groundwork for future efforts to understand the mechanisms by which malaria vector mosquitoes adapt to a heterogeneous and changing environment.
In the An. gambiae complex, three species (An. merus, An. melas, and An. bwambae) are tolerant of brackish water, unlike the other members of the group that are obligate freshwater breeders. Previously, we worked with the only laboratory-colonized salt tolerant species, An. merus, and conducted classical QTL mapping studies as well as transcriptomics to identify candidate loci. However, our understanding of the genetic basis of the physiological adaptation to breeding in saltwater would be greatly enhanced by knowledge of the evolution of this trait across species in the An. gambiae complex. Did it arise independently in the three salt tolerant species, as surmised by Coluzzi et al (1979)?
Understanding the evolution of this adaptive trait is contingent on a phylogenetic framework, yet the previous phylogenomic study of the An. gambiae complex by Fontaine et al (2015) did not incorporate An. bwambae. This species is highly endemic, apparently limited to the hot springs of the Semliki Forest in western Uganda, and it has not been colonized (since historical times). Thus, to address these questions, we began by collecting An. bwambae and performed individual whole genome sequencing with the dual goals of constructing a reference genome assembly, and generating SNP data that could guide reconstruction of the population genomic history of An. bwambae. With the reference assembly completed and aligned to the other reference assemblies for the An. gambiae complex, analyses of species phylogeny, introgression, and population genomics are underway. In the process, we believe we have found a polymorphic inversion in An. bwambae on the X-chromosome. We plan to focus particular attention on those genomic regions whose phylogeny groups all three salt tolerant species together (in contradiction to the species tree).
Funding:
Support for this work comes from Target Malaria, which receives core funding from the Bill and Melinda Gates Foundation and from the Open Philanthropy Project Fund, an advised fund of Silicon Valley Community Foundation.
References:
- Uyhelji HA, Cheng C, Besansky NJ. (2016) Transcriptomic differences between euryhaline and stenohaline malaria vector sibling species in response to salinity stress. Mol Ecol 25: 2210-25.
- Smith HA, White BJ, Kundert P, Cheng C, Romero-Severson J, Andolfatto P, Besansky NJ. (2015) Genome-wide QTL mapping of saltwater tolerance in sibling species of Anopheles (malaria vector) mosquitoes. Heredity 115: 471-479.
- White BJ, Kundert PN, Turissini DA, Van Ekeris L, Linser PJ, Besansky NJ. (2013) Dose and developmental responses of Anopheles merus larvae to salinity. J Exp Biol 216: 3433-3441.
Collaborators (Past and Present):
- University of Notre Dame Frédéric Labbé, Becca Love, Michael Fontaine, Christina Bergey, Bradley White, Peter Kundert, Changde Cheng, Jeanne Romero-Severson, Hilary (Smith) Uyhelji, Scott Small, Nora Besansky
- Princeton University: Peter Andolfatto
- University of California, Riverside: Bradley White, David Turissini
- University of Florida: Leslie Van Ekeris, Paul Linser
- University of Groningen: Jorge Eduardo Amaya-Romero, Michael Fontaine
- Liverpool School of Tropical Medicine and Hygiene: Harold Townson
- University of Lausanne, Switzerland: Rob Waterhouse
- Uganda Viral Research Institute: Jonathan Kayondo and his team
- Max Planck Institute for Evolutionary Biology, Germany: Julien Y Dutheil
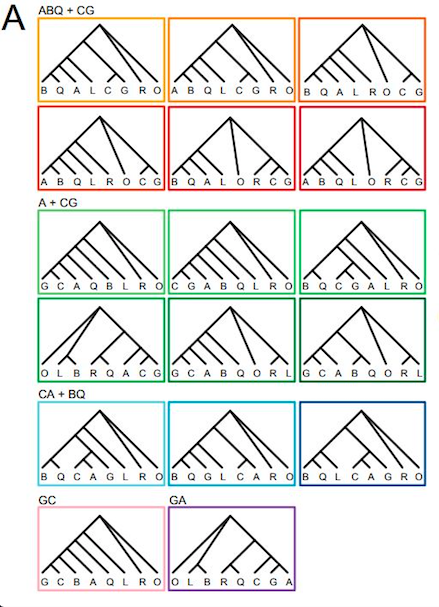
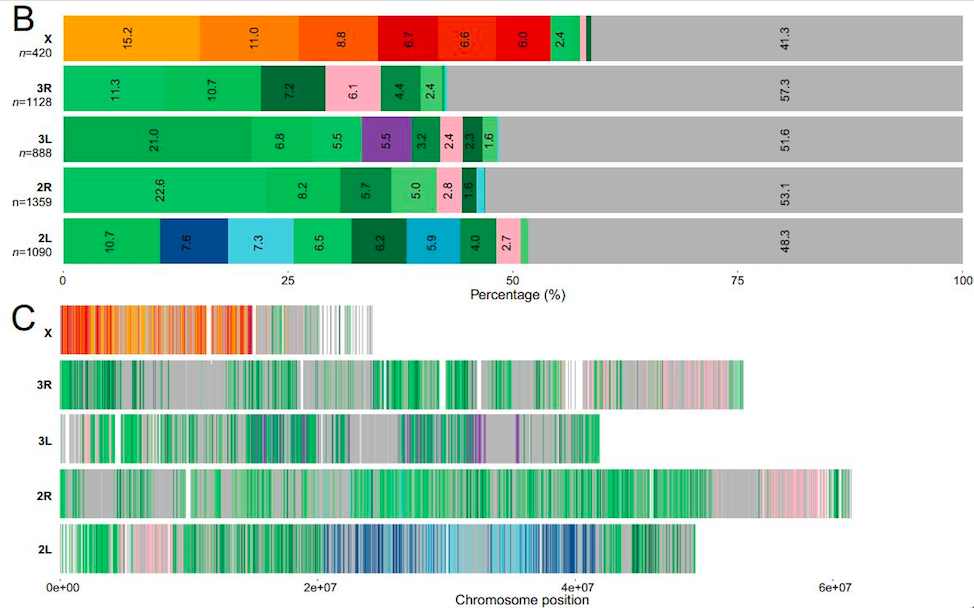 Phylogenetic relationships in the An. gambiae complex. A: The 17 most common ML-rooted phylogenies across the genome (>5% at least for one chromosome), inferred from n=4885 50-kb genomic windows for An. arabiensis (A), An. coluzzii (C), An. gambiae (G), An. melas (L), An. merus (R), An. bwambae (B), An. quadriannulatus (Q), and An. christyi as out-group (O). These topologies are painted as correspondingly colored blocks on chromosome arms, according to their frequency in panel B, or by window in panel C. Topologies specific to 2La are represented in blue on 2L; those specific to 3La are represented in purple on 3L. Gray represents all other observed topologies pooled.
Phylogenetic relationships in the An. gambiae complex. A: The 17 most common ML-rooted phylogenies across the genome (>5% at least for one chromosome), inferred from n=4885 50-kb genomic windows for An. arabiensis (A), An. coluzzii (C), An. gambiae (G), An. melas (L), An. merus (R), An. bwambae (B), An. quadriannulatus (Q), and An. christyi as out-group (O). These topologies are painted as correspondingly colored blocks on chromosome arms, according to their frequency in panel B, or by window in panel C. Topologies specific to 2La are represented in blue on 2L; those specific to 3La are represented in purple on 3L. Gray represents all other observed topologies pooled.