Milenkovic Lab
| ABOUT | DOCUMENTATION AND TUTORIAL | DOWNLOAD DynaMAGNA++ | DOWNLOAD DATA |
Section 1: System Requirements
DynaMAGNA++ runs on Linux, Mac OS X, and Windows, and is available for download as a compressed file here. Statically compiled executables are provided for all three operating systems, so the aligner is ready to run immediately after downloading, though instructions are also provided to compile directly from the source.
The suggested network size for DynaMAGNA++ is up to 2,000 nodes and 100,000 events. The aligner's memory requirement increases linearly with respect to the number of nodes and events. The time taken to run the aligner increases linearly with respect to the number of events, with the running time decreasing close to linearly with the number of cores used.
The DynaMAGNA++ download takes up 1-2MB of disk space, though more space will be required to store networks and the resulting alignments between them.
Installing involves simply uncompressing the downloaded file and running DynaMAGNA++.
Section 2: Using DynaMAGNA++
2.1: Graphical User Interface (GUI)
Start DynaMAGNA++ by running the executable. DynaMAGNA++ comes with a user-friendly graphical interface; below is a screenshot of the interface along with descriptions of each input field.
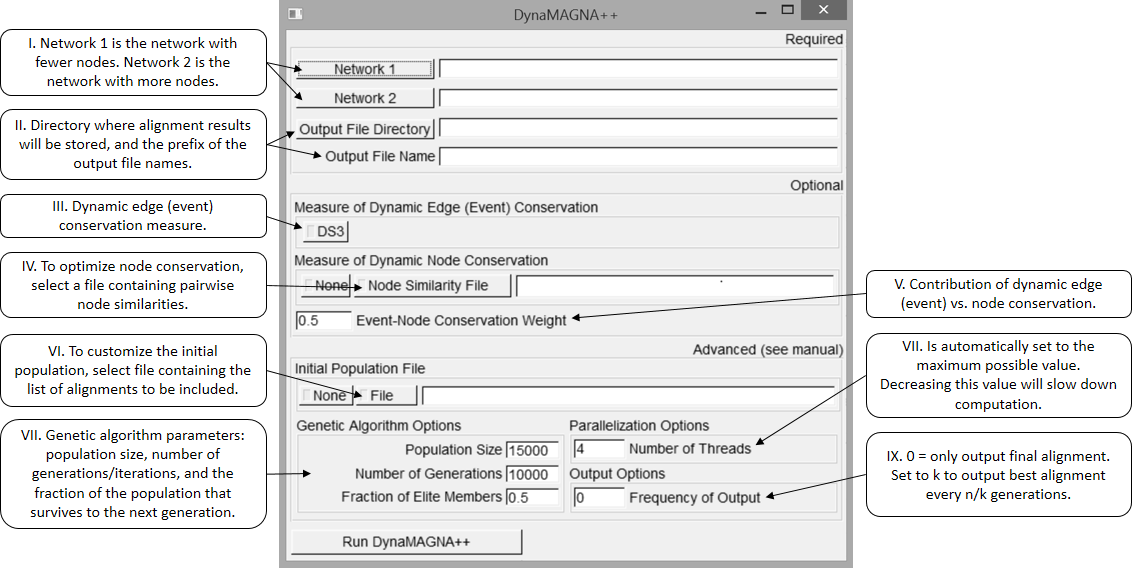
I.
Choose two networks to align. Network 1 must have fewer nodes than Network 2. DynaMAGNA++ accepts networks in the event list format (see an example). Toy example networks can be found here.
II.
Choose the output directory and a prefix for each output file name. For example, if "ex" is entered, then "ex" will appear at the beginning of the name of each file that DynaMAGNA++ outputs. DynaMAGNA++ will output files for the resulting alignments, the parameters used to create the alignments, and some statistics of the alignments. See Section 3 for a more detailed description of output files.
III.
Choose an dynamic edge (event) conservation measure to optimize. DS3 is currently the only measure implemented..
IV.
Choose whether to optimize a node-based measure in addition to optimizing an event-based measure. If "None" is selected, then DynaMAGNA++ will only optimize edge conservation (see IV). If "Node Similarity File" is selected, then choose a node similarity file. DynaMAGNA++ can read node similarity files in two formats: the node pairs format (see an example) or the CSV matrix format (see an example).
Supposing we align Network 1 and Network 2. The edge list or CSV matrix format should have the first column containing the nodes in Network 1, the second the nodes in Network 2, and the third column the similarity value between the two nodes.
V.
Choose a parameter ![]() . Using this parameter, DynaMAGNA++ can simultaneously optimize an event-based measure (DS3) and a node-based measure (of topological or biological node similarities). In particular, given an event-based measure
. Using this parameter, DynaMAGNA++ can simultaneously optimize an event-based measure (DS3) and a node-based measure (of topological or biological node similarities). In particular, given an event-based measure ![]() , a node-based measure
, a node-based measure ![]() , and a parameter
, and a parameter ![]() between 0 and 1, DynaMAGNA++ will optimize
between 0 and 1, DynaMAGNA++ will optimize ![]() . When
. When ![]() is 1, DynaMAGNA++ will only optimize event conservation, and when
is 1, DynaMAGNA++ will only optimize event conservation, and when ![]() , DynaMAGNA++ will only optimize node conservation.
, DynaMAGNA++ will only optimize node conservation.
VI.
DynaMAGNA++ can either improve an
existing set of alignments or start from
scratch. Choose either "None" or "File". If "File"
is selected, then DynaMAGNA++ will prompt for an
initial population file, which will define an
existing set of alignments for which to
improve. This file should contain a list of
alignments file names (see
an example), which
must be in the same directory as the initial
population file. If "None" is selected, no initial
population will be used, so DynaMAGNA++ will start
from scratch by generating a random set of initial
alignments.
VII.
DynaMAGNA++ uses a genetic algorithm, for which there are three parameters:
"Population Size" is the population size of the genetic algorithm; that is, the genetic algorithm will work with this many alignments with each iteration. If no initial population is provided (see VI), then 15,000 is generally the recommended population size.
"Number of Generations" is the number of iterations or generations for which the genetic algorithm will run. If no initial population is used, then 10,000 is generally the recommended number of generations. If using an alignments generated by another algorithm, then 1,000 is the recommended number of generations.
"Fraction of Elite Members" is the highest-quality fraction of the population that survives to the next generation. 0.5 is always the recommended value.
VIII.
DynaMAGNA++ is capable of using multiple threads to perform its computation. It parallelizes the calculation of alignment quality, which is the computational bottleneck of DynaMAGNA++. Calculation of alignment quality is divided among threads. DynaMAGNA++ automatically determines the number of threads by using as many threads as there are cores in the computer, though this value can be changed manually. However, using more threads than there are cores is not recommended since performance is not likely to improve. Performance will drop if too many threads are used.
IX.
Choose how often DynaMAGNA++ outputs the best alignment in the population. If the frequency of output is k, and there are n generations, then DynaMAGNA++ will output the best alignment every n/k generations. For example, if k = 1, then DynaMAGNA++ will output the best alignment with every generation, and if k = n, then DynaMAGNA++ will only output the best alignment of the last generation. It is recommended that k divides n.
2.2: Command Line Interface (CLI)
Instructions for running the command line interface (CLI) of DynaMAGNA++ can be found in the README.txt file that accompanies the CLI executables of DynaMAGNA++.
Section 3: Output and Results
Upon beginning an execution (and supposing the prefix defined in 2.II is "ex_"), DynaMAGNA++ generates three output files by default. The final alignment file "ex_final_alignment.txt" consists of the aligned pairs of nodes calculated by DynaMAGNA++. The alignment statistics file "ex_final_stats.txt" contains the DS3, node similarity, and the overall score of the final alignment. The common conserved subgraph file, "ex_final_visualize.sif", contains the common conserved subgraph of the alignment of the two networks. It provides an intuitive visualization of the final alignment, and can be viewed using a program such as Cytoscape.
If the frequency of output option is set to k, then DynaMAGNA++ outputs the details of the best alignment every n/k generations (where n is the total number of generations, asdefined in 2.XI). Each alignment is written to a file, containing a list of node pairs. Nodes in the left column are from Network 1 and the nodes on the right are from Network 2. Also according to the frequency of output, DynaMAGNA++ will append statistics of the best alignment to the file "ex_stats.txt".
The format of the output alignment file names is "ex_M_p_n_i.aln", where M is the edge conservation measure (one of S3), p is the population size, n is the total number of generations, and i is the generation that the alignment was written. The alignment from the last generation (ex_M_p_n_n.aln) is the final alignment produced by DynaMAGNA++.
DynaMAGNA++ will also create a file "ex_params.txt", which contains a summary of the initial parameters given.
Section 4: Authors, Credits, and Licensing
DynaMAGNA++ is created by V. Vijayan, V. Saraph and T. Milenkovic at University of Notre Dame.
DynaMAGNA++ is freely available for academic use only.
Disclaimer: THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.