Structural Genomics
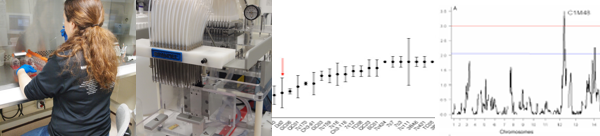
This segment of the lab grew from my interest in the adaptive role of copy number variation and my research group’s embrace of emerging genomic technologies. We developed a gene chip for comparative genome hybridization (CGH) and single nucleotide polymorphism (SNP) genotyping (An optimized microarray platform for assaying genomic variation in Plasmodium falciparum field populations, Genome Biol, 2011). (Adaptive copy number evolution in malaria parasites, PLoS Genet, 2008; The landscape of inherited and de novo copy number variants in a Plasmodium falciparum genetic cross, BMC Genomics, 2011). Furthermore, we used a new sequencing technology to explore inherited structural variation (High-throughput 454 resequencing for allele discovery and recombination mapping in Plasmodium falciparum, BMC Genomics, 2011). These data improved the malaria linkage map and its relationship to the physical genome (Array-based high-throughput marker discovery for integrating genetic and physical maps in Plasmodium falciparum, pending revision), and are partnered with the Sanger Center to sequence all the progeny of 3 malaria crosses.